diff --git a/content/100.figure-table-legends.md b/content/100.figure-table-legends.md
index 86ccfb6..3d75225 100644
--- a/content/100.figure-table-legends.md
+++ b/content/100.figure-table-legends.md
@@ -1,7 +1,7 @@
## Figure Titles and Legends {.page_break_before}
-{#fig:fig1 tag="1" width="7in"}
+{#fig:fig1 tag="1" width="7in"}
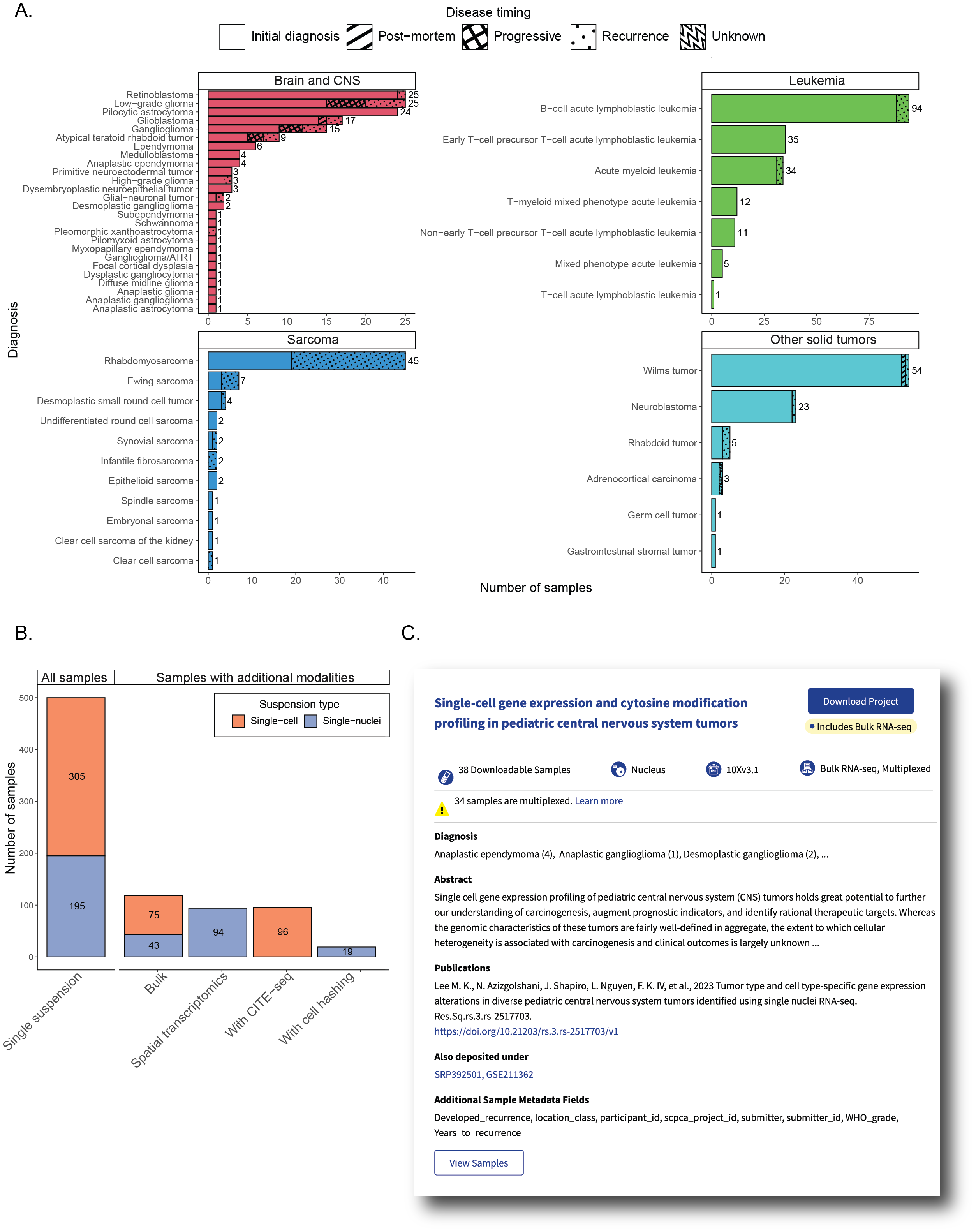
A. Barplots showing sample counts across four main cancer groupings in the ScPCA Portal, with each bar displaying the number of samples for each cancer type.
Each bar is shaded based on the number of samples with each disease timing, and total sample counts for each cancer type are shown to the right of each bar.
@@ -19,7 +19,7 @@ Where available, submitter-provided citation information, as well as other datab
-{#fig:fig2 tag="2" width="7in"}
+{#fig:fig2 tag="2" width="7in"}
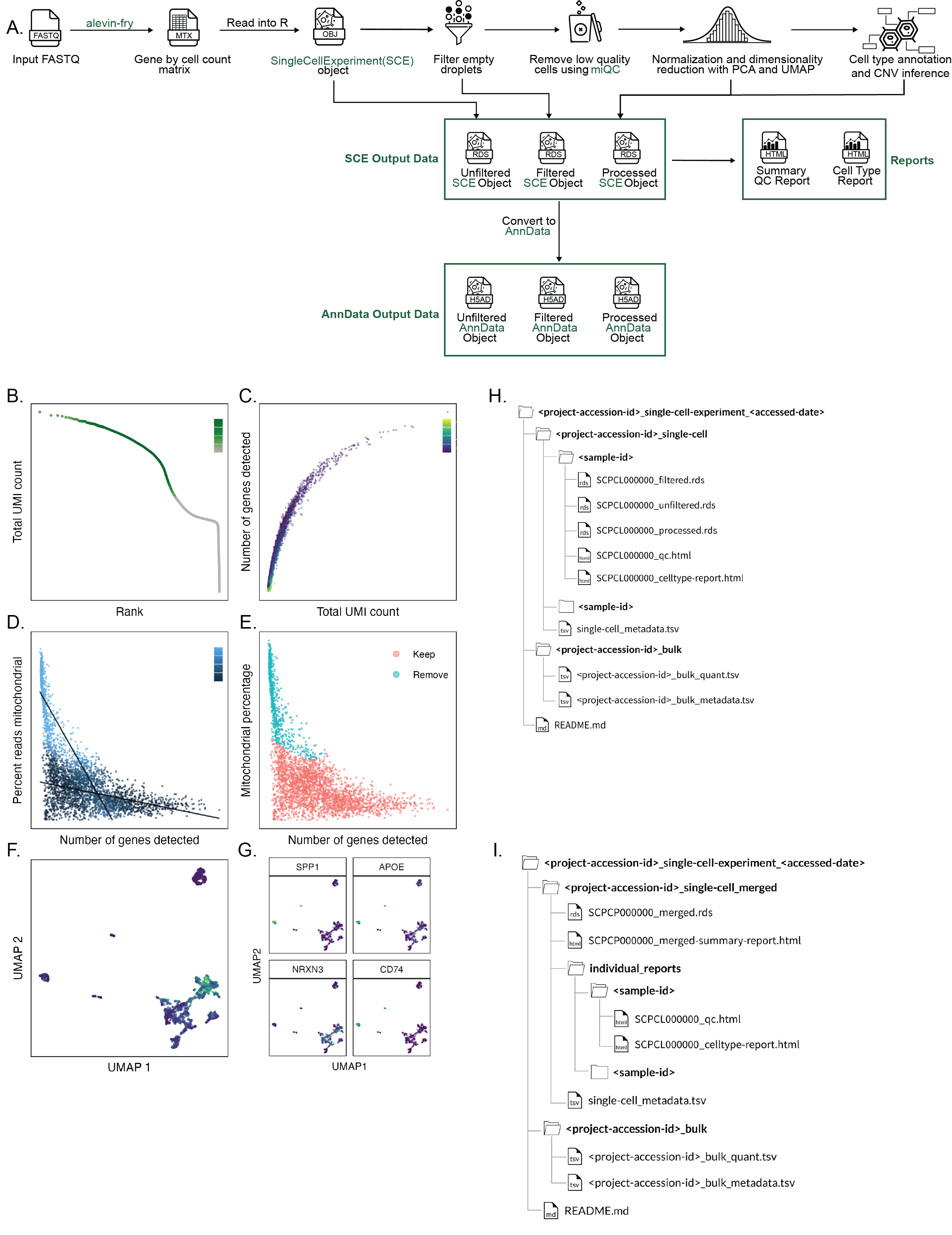
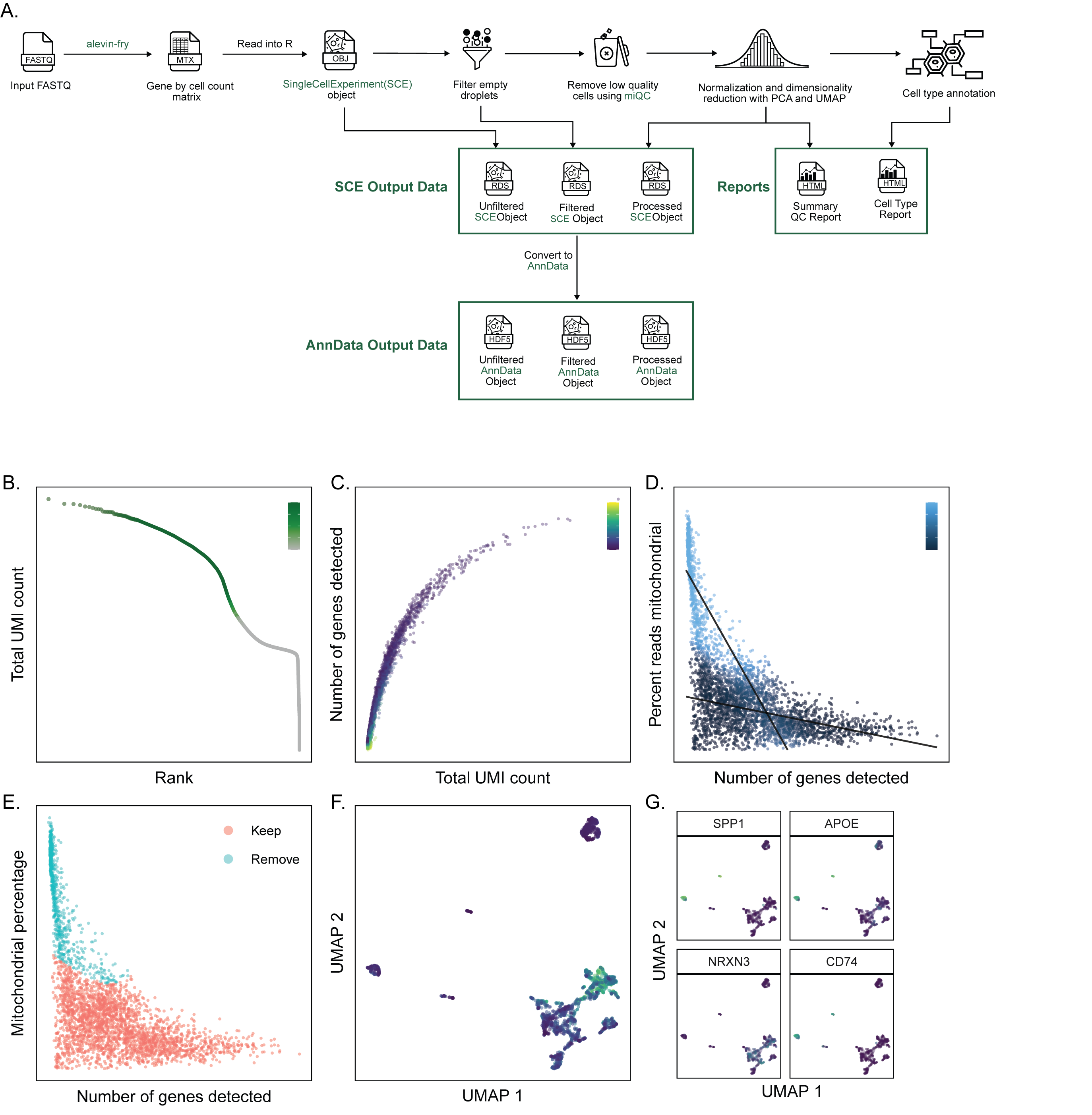
A. Overview of `scpca-nf`, the primary workflow for processing single-cell and single-nuclei RNA-seq data for the ScPCA Portal.
Mapping is first performed with `alevin-fry` to generate a gene by cell count matrix, which is read into `R` and converted into a `SingleCellExperiment` (`SCE`) object.
@@ -50,7 +50,7 @@ In the actual summary QC report, the top 12 most highly variable genes are shown
-{#fig:fig3 tag="3" width="7in"}
+{#fig:fig3 tag="3" width="7in"}
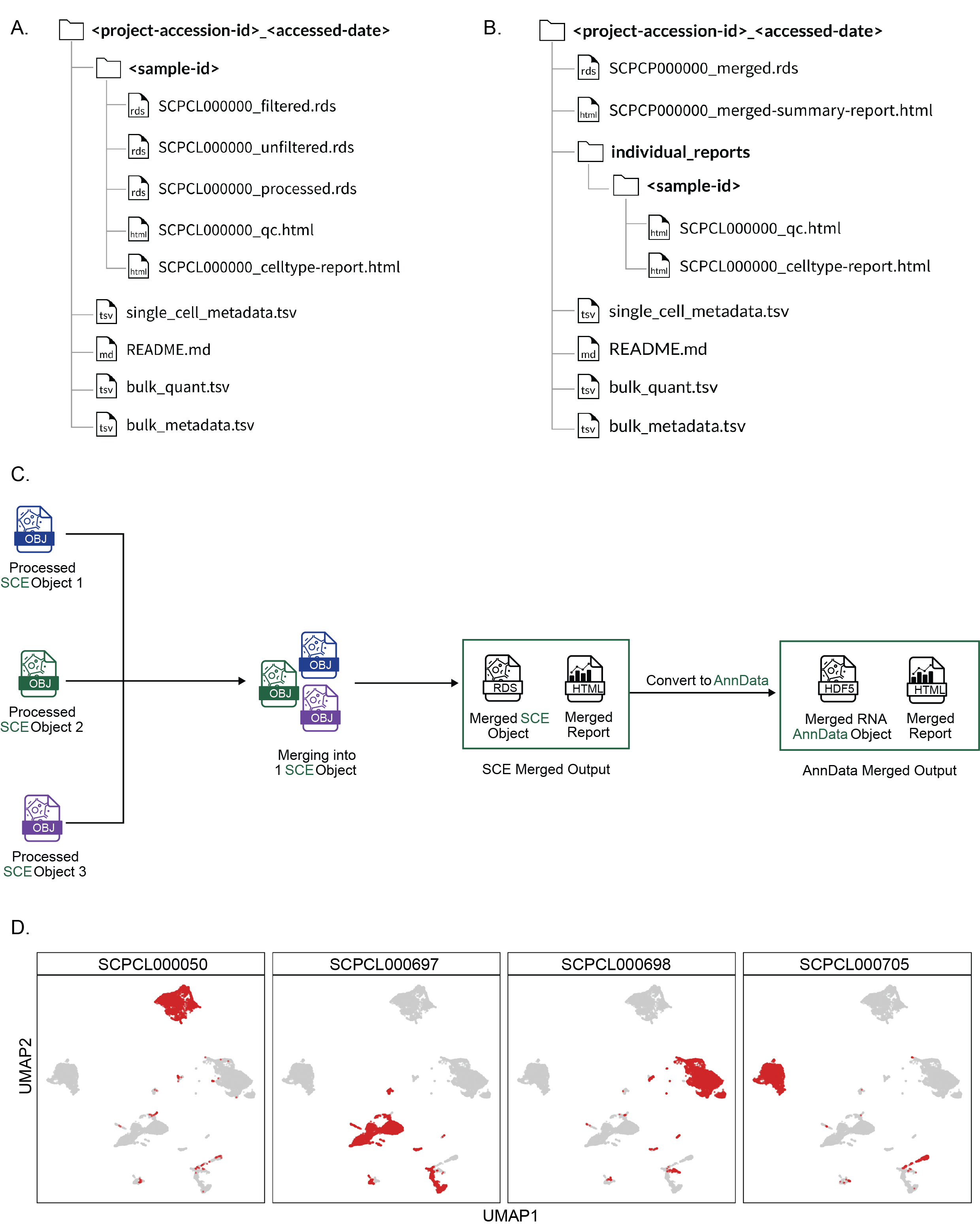
A. File download structure for an ScPCA Portal project download in `SingleCellExperiment` (`SCE`) format.
The download folder is named according to both the project ID and the date it was downloaded.
@@ -76,7 +76,7 @@ The libraries pictured are a subset of libraries in the ScPCA project `SCPCP0000
-{#fig:fig4 tag="4" width="7in"}
+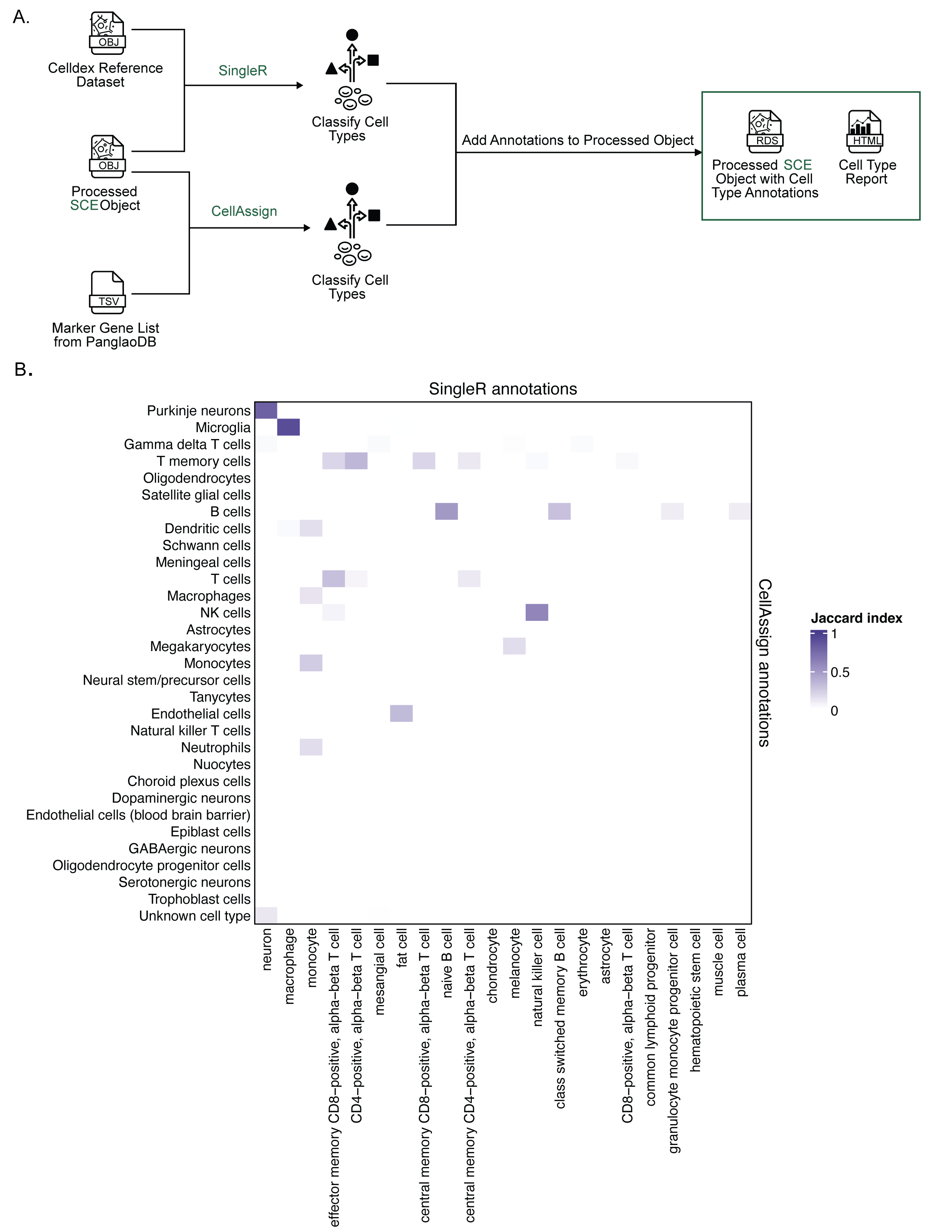{#fig:fig4 tag="4" width="7in"}
A. Expanded view of the process for adding cell type annotations within `scpca-nf`, as introduced in Figure {@fig:fig2}A.
Cell type annotation is performed on the `Processed SCE Object`.
@@ -117,7 +117,7 @@ The reference includes marker genes for all cell types present in each organ.
-{#fig:figS1 tag="S1" width="7in"}
+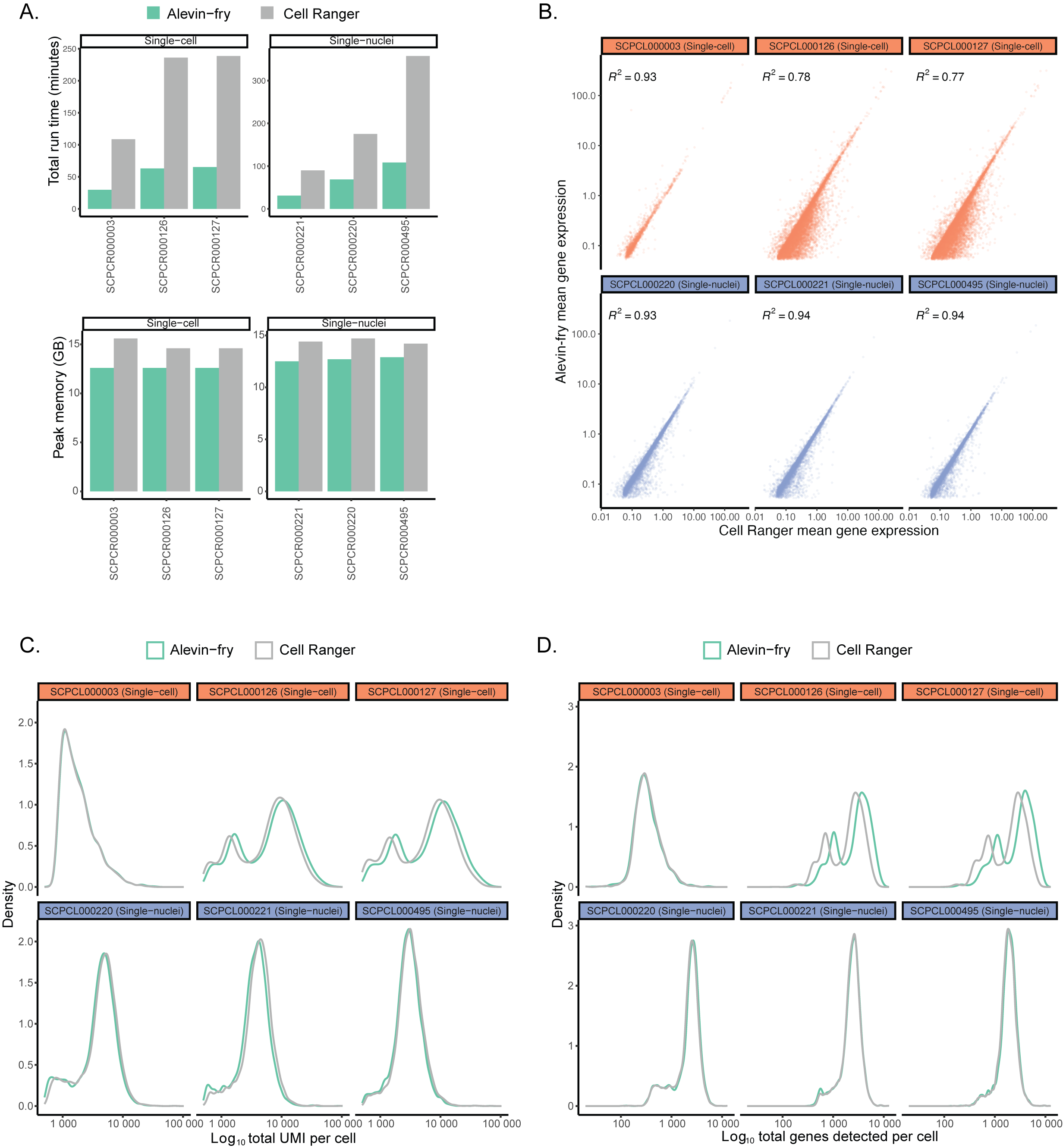{#fig:figS1 tag="S1" width="7in"}
Each panel compares metrics for six representative ScPCA libraries, including three single-cell and three single-nuclei suspensions, obtained from processing libraries with both `alevin-fry` and `CellRanger`.
@@ -138,7 +138,7 @@ Distributions reflect broad agreement between platforms in the total number of g
-{#fig:figS2 tag="S2" width="7in"}
+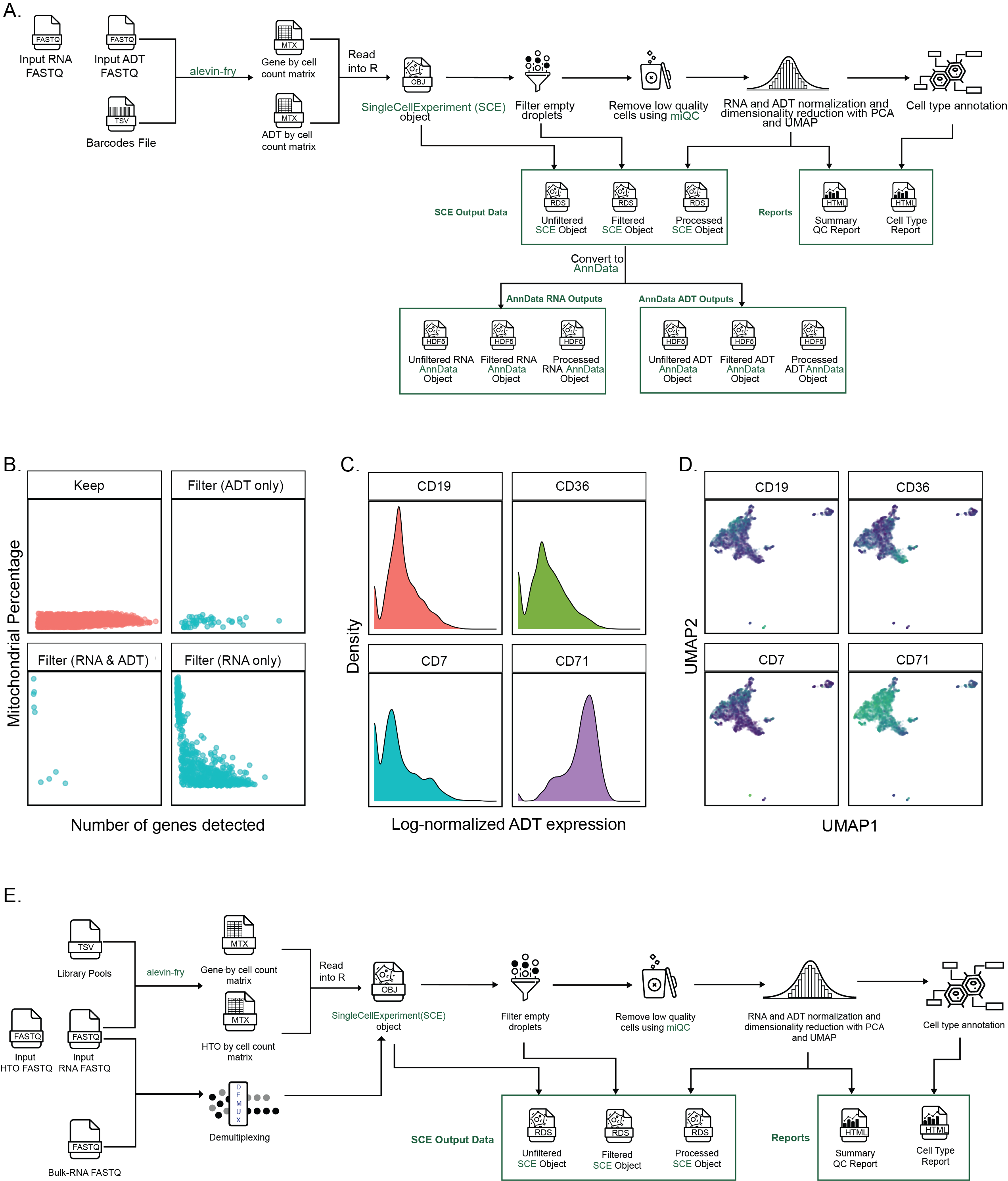{#fig:figS2 tag="S2" width="7in"}
A. Overview of the `scpca-nf` workflow for processing libraries with CITE-seq or antibody-derived tag (ADT) derived data.
The workflow mirrors that shown in Figure {@fig:fig2}A with several differences accounting for the presence of ADT data.
@@ -172,7 +172,7 @@ Third, only `SCE` files are provided for multiplexed libraries; no corresponding
-{#fig:figS3 tag="S3" width="7in"}
+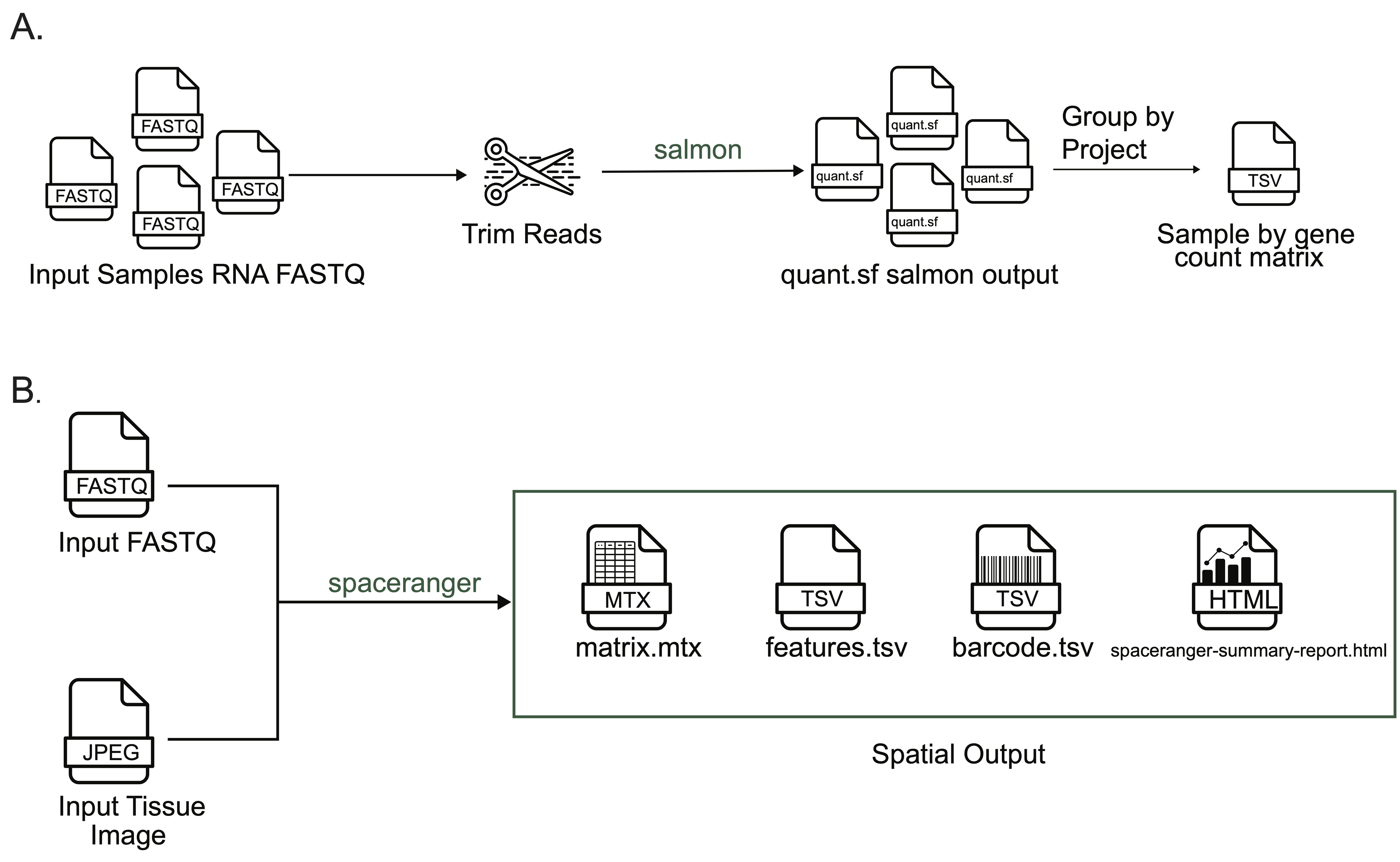{#fig:figS3 tag="S3" width="7in"}
A. Overview of the bulk RNA-Seq workflow.
A set of FASTQ files from libraries sequenced with bulk RNA-seq are provided as input.
@@ -185,7 +185,7 @@ The workflow directly returns the results from running `spaceranger` without any
-{#fig:figS4 tag="S4" width="7in"}
+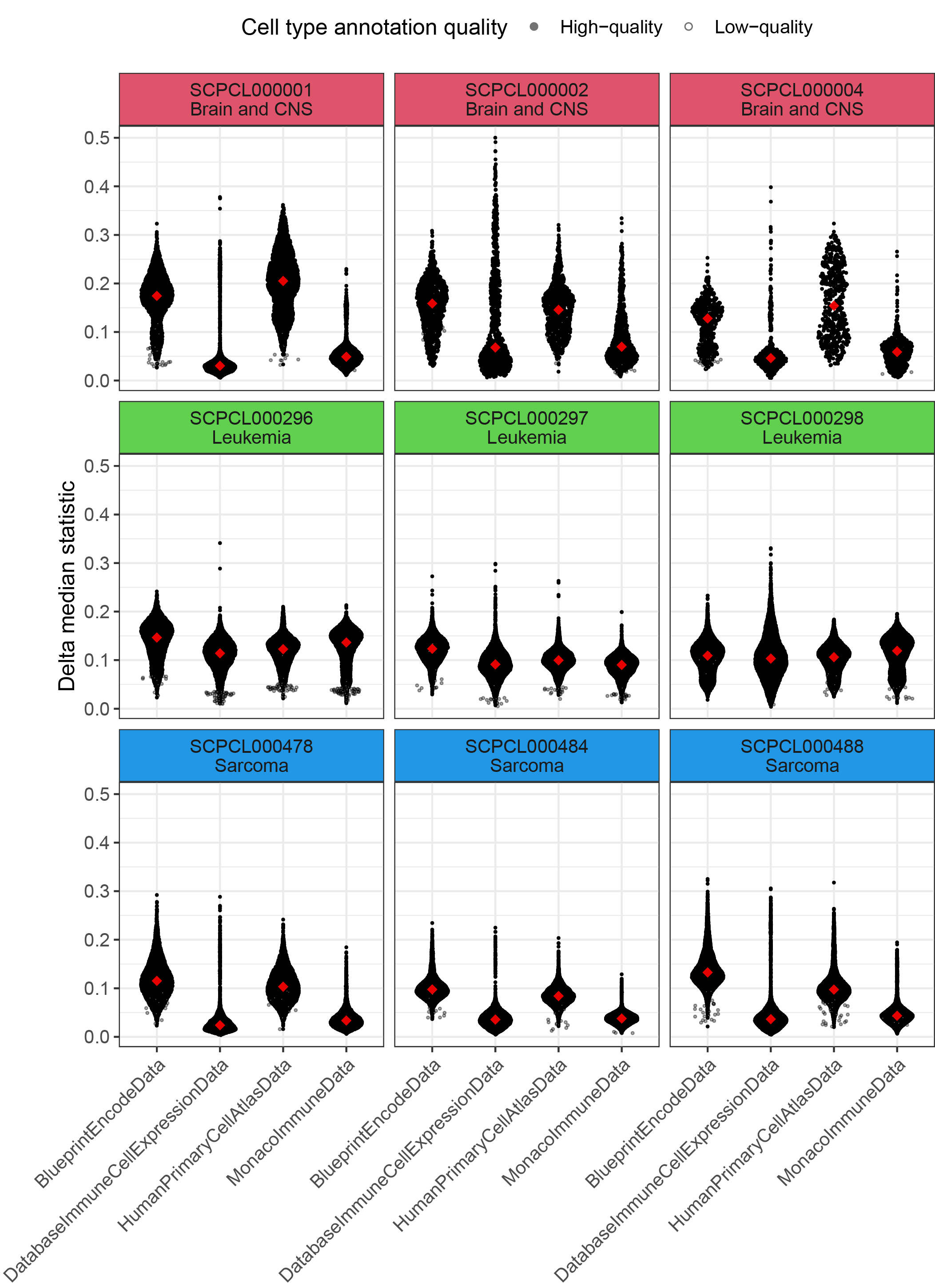{#fig:figS4 tag="S4" width="7in"}
`SingleR` was used to annotate ScPCA libraries using four different human-specific references from the `celldex` package.
Libraries represent three different diagnosis groups in the ScPCA Portal - Brain and CNS, Leukemia, and Sarcoma - as indicated in the labels for the individual panels.
@@ -196,7 +196,7 @@ Red diamonds represent the median delta median score for all cells with high-qua
-{#fig:figS5 tag="S5" width="7in"}
+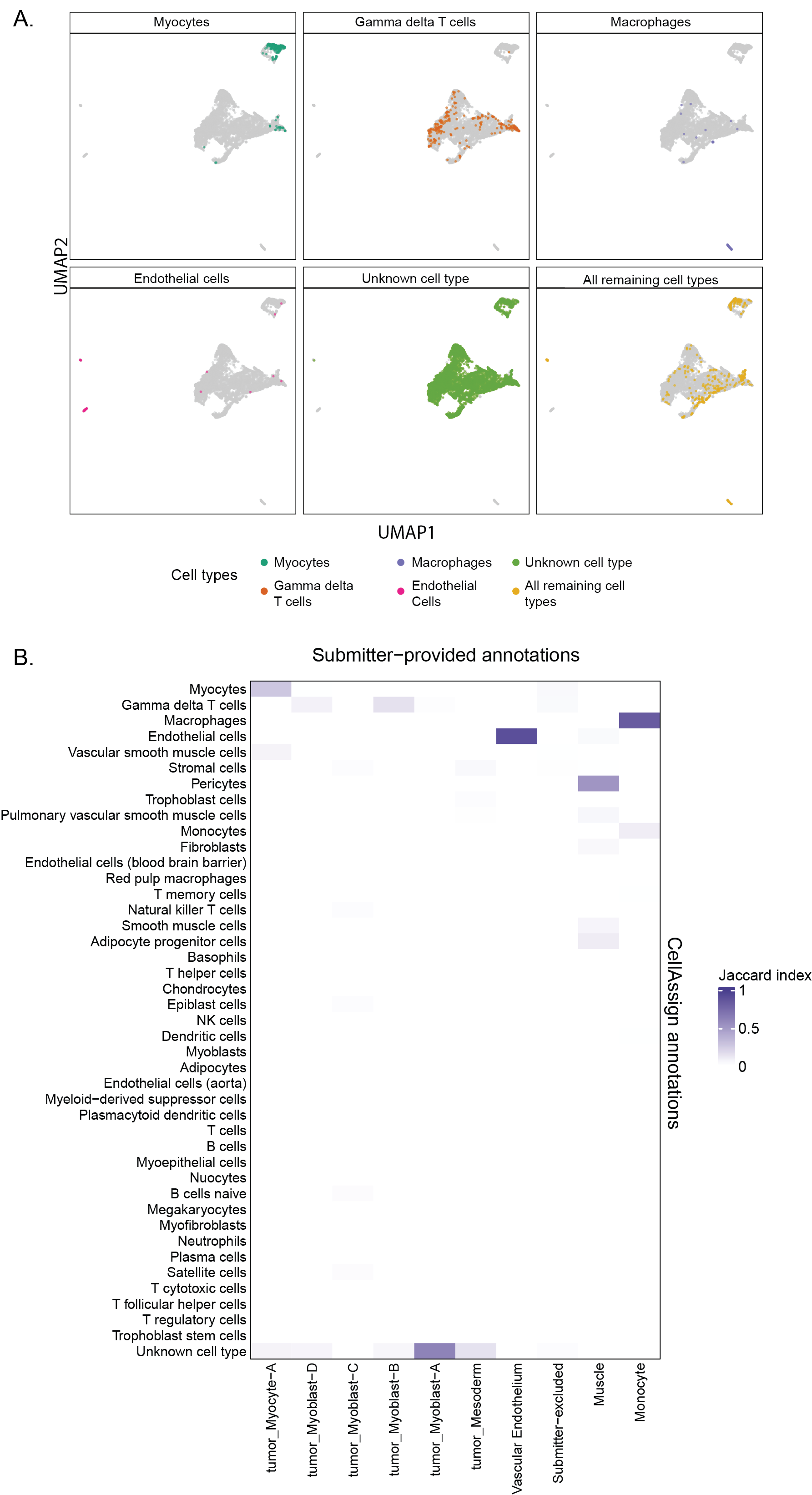{#fig:figS5 tag="S5" width="7in"}
Both plots in this figure are examples of plots that display results from annotating cells with `CellAssign` that can be found in the cell type summary report, shown here for library `SCPCL000490`[@doi:10.1016/j.devcel.2022.04.003].
@@ -209,7 +209,7 @@ A value of 1 means that there is complete overlap between which cells are annota
-{#fig:figS6 tag="S6" width="7in"}
+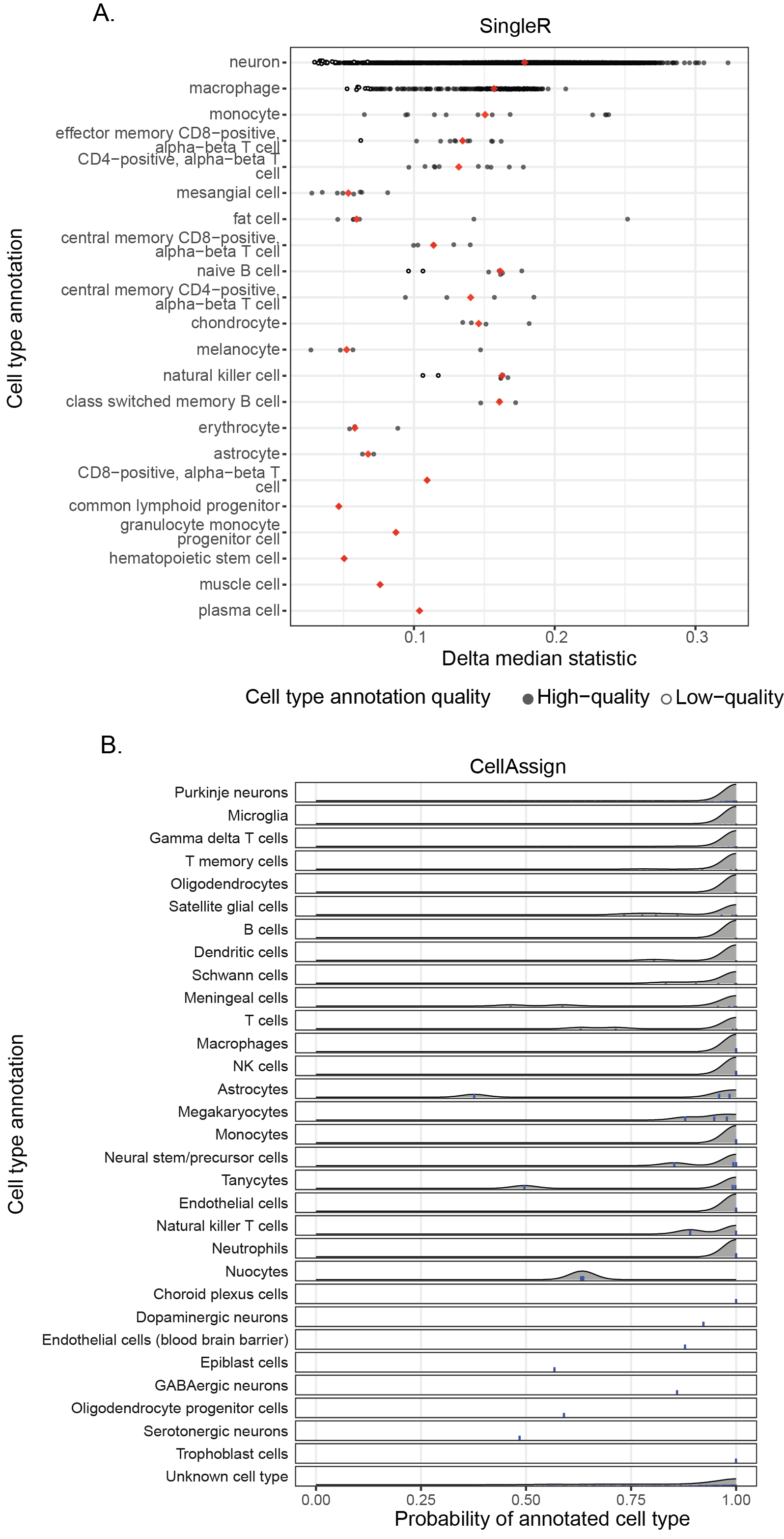{#fig:figS6 tag="S6" width="7in"}
Both plots in this figure are examples of diagnostic plots in the cell type summary report, shown for library `SCPCL000001` [@doi:10.1093/neuonc/noad207].
@@ -227,7 +227,7 @@ Taller line segments are shown for any distribution with five or fewer cells.
-{#fig:figS7 tag="S7" width="7in"}
+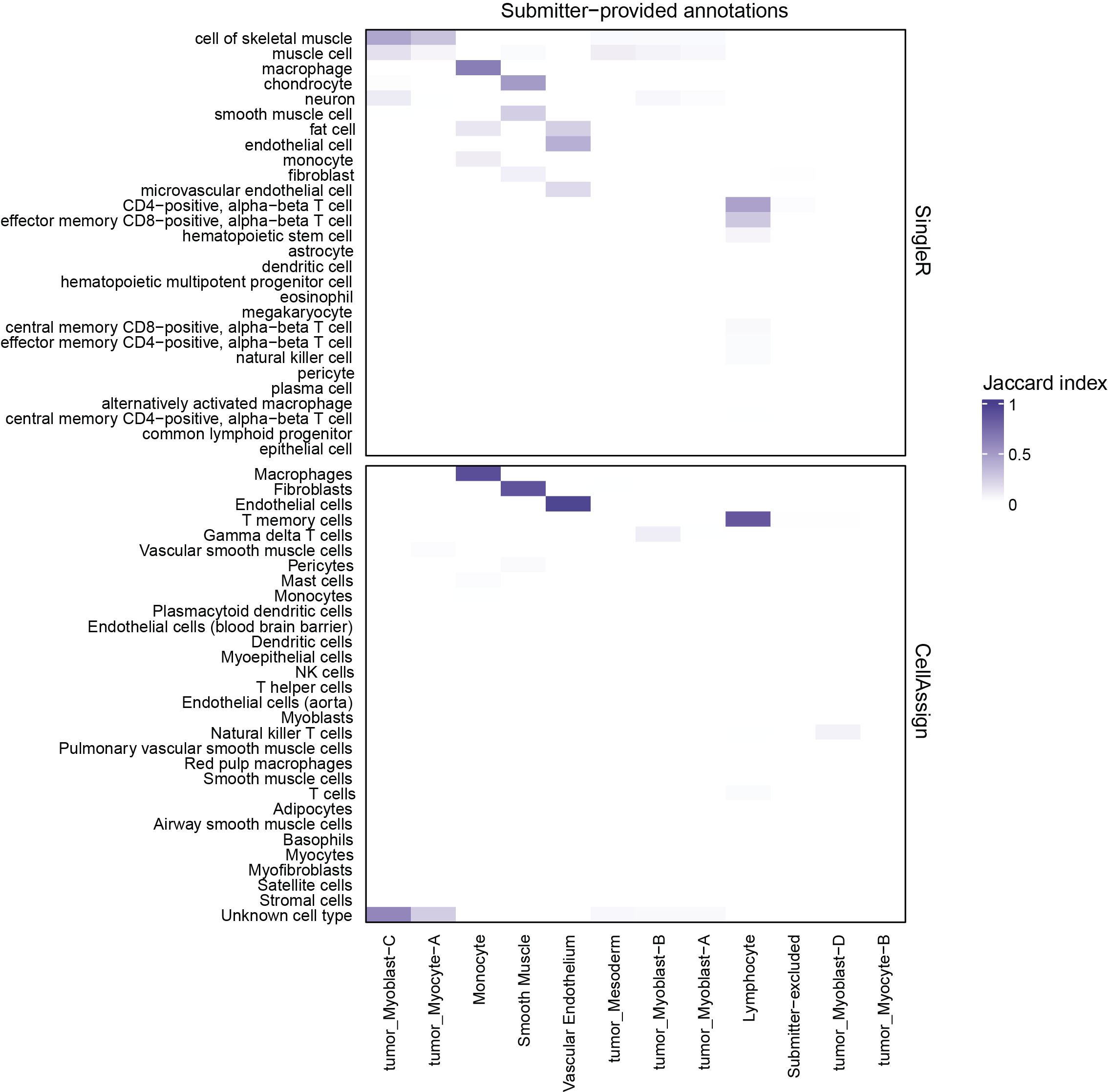{#fig:figS7 tag="S7" width="7in"}
This example heatmap from the cell type summary report compares submitter-provided annotations to annotations with `SingleR` and `CellAssign`, shown for library `SCPCL000498` [@doi:10.1016/j.devcel.2022.04.003].
This heatmap is only shown in the cell type summary report if submitters provided cell type annotations.