| title | author | date | output | editor_options | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Reproducibel research, part I |
03 Sep 2024 |
|
|
-
DataCamp, I will assign the courses on RMarkdown and git. ON DataCamp
-
Read Good enough practices in scientific computing by Wilson et al.
-
Read chapters 1 and 2 in Bioinformatics Data Skills
-
An alternative reading on version control and using git and github is [chapter 2 in the CSB] book(https://doi-org.libaccess.lib.mcmaster.ca/10.1515/9780691183961-006). Pick which one you prefer.
- Is publishing your paper enough?
- Data?
- Meta-data?
- Analysis scripts/code?
What easy steps can make science more reproducible & helpful for future scientists?
- You want to make this process as easy as possible for yourself first and foremost.
- In the process of organizing your data and analysis pipelines to share, you will make yourself a better scientist.
- Future you (6 months after you finish data collecting), will thank you too!
"Grant recipients are required to deposit into a recognized digital repository all digital research data, metadata and code that directly support the research… The repository will ensure safe storage, preservation, and curation of the data. The agencies encourage researchers to provide access to the data ... Whenever possible, these data, metadata and code should be linked to the publication with a persistent" See links here
-
At the onset of your research, organize your workflow with the goal of making it as easy as possible to share.
-
With a few simple guidelines, you will save yourself an enormous amount of time while making your data and your analysis scripts available.
/ProjectName
./data
./scripts
./outputs
./misc
./manuscript
READMEAll projects should have very similar organizational formats:
-
Informative project name.
-
datafolder contains raw data as a flat file (tabular data, comma seperated), database or if necessary a proprietary spreadsheet. -
scriptsfolder contains scripts/code you use to analyse your data. -
outputsfor automated outputs (tables and figures from the scripts + data) -
miscfor miscellaneous. Additional meta-data for files, experimental design information, readme of overall project. Figures that can not easily be generated.
-
File formats for many commercial programs (SAS, JMP, Excel,) can not be easily read by other programs.
-
Worse still. As new versions of software emerge, no guarantee of being able to open older file formats (Excel). -- It is fine to use Excel. But when sharing data (and scripts) use formats like .txt (such as tab delimited or .tsv) or .csv (comma seperated) which can be easily read.
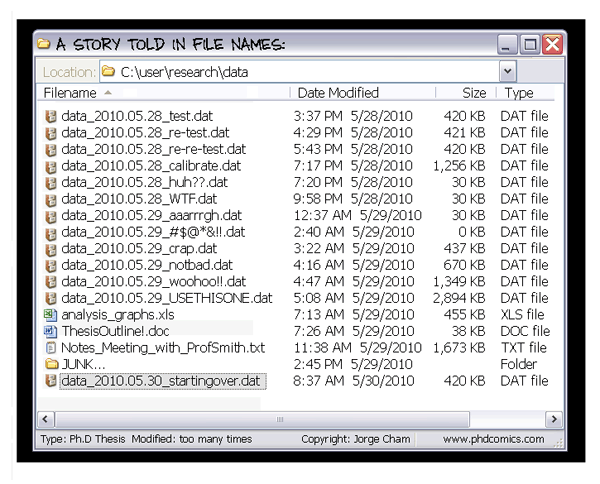
- Many projects generate hundreds or thousands of files. For instance digital microscopy, automated measuring tools, sequencing, etc.
- Spend a few minutes thinking about how to name your files. This will save you hours or days down the road!
Some "do's" and "do not"
DO: Use the same naming conventions for every file. i.e.
ID_Dmel_M_HN_01.tif
ID_Dmel_M_HN_02.tif
ID_Dmel_M_HN_03.tif
ID_Dmel_F_HN_01.tif
ID_Dmel_F_HN_02.tif
ID_Dmel_F_HN_03.tif
DO NOT:Use spaces in file names. Use consistent delimiters, and limit it to one type of delimiter if you can. i.e. underscores!
ID_Dmel_M_HN_01.tif
Not ID Dmel_M_HN 01.tif
Not ID_Dmel-M_HN.01.tif
DO: Use consistent delimiters! Underscore _ and dash - are good choices.
DO: Try to have as much of the experimental information in the file name (like below)
ID_Dmel_M_HN_01.tif
ID = initials
Dmel = species name
M = sex (M/F)
HN = High Nutrition
01 = specimen number
Why is this important?
It allows for all of the experimental variables to be extracted in an automated way (even in Excel).
But having inconsistencies in naming can make a 1 minute activity or one line of code into something not fun.
At least 40% of my coding time is spent parsing and cleaning badly organized data, in particular due to naming conventions.
-
Version control is about how to manage and keep track of editing and revisions on any kind of document. This can be code for software, your analysis pipeline, or manuscript editing.
-
The basic idea is that without keeping track of edits and revisions you make can be problematic.
-
version control keeps track of changes you make to files (differences), when you made them. It also forces you to make a small comment associated with each change to help you find when you made a particular change.
-
This also enables collaborations with other people (with respect to revising code or documents), as you can see what changes they made, and can choose to accept them (or not).
-
There are many version control systems out there.
-
Don’t assume that any collaborators or future users (or future you) will know what variables are in your data set.
-
Write a short readme file containing basic meta-data for the project including: explanations of the variables, how and where they were collected (and why), file naming conventions etc.
-
If you do this while organizing everything it takes just a few minutes, but it will save future you and collaborators so much time.
In addition to backing up data for yourself, getting it out there as soon as possible (in conjunction with manuscript submission or acceptance) is crucial.
-
There are often both institutional resources for long term data archiving, and broader initiatives.
-
Some of the Data sharing portals are fairly general purpose (DRYAD, zenodo, figshare, dataverse, FRDR in Canada).
-
Some are highly specialized for certain data types (Morphobank, tree of life, NCBI GEO,NCBI SRA, morphosource, DigiMorph