Prepared by Zhe Feng ([email protected])
Pacific Northwest National Laboratory
All tracking parameters are set in a config file (config.yml). Each tracking step produces netCDF file(s) as output and can be run separately if consistent output netCDF files from previous steps are available. This design allows certain time-consuming steps to be run in parallel and only need to be only once. For example, once feature identification and consecutive linking in Step 1 and 2 (see Section 2.2) are produced during a period, tracking during any sub-periods only requires running Step 3 and subsequent steps.
PyFLEXTRKR works with netCDF files using Xarray's capability to handle N-dimension arrays of gridded data. Currently, PyFLEXTRKR supports tracking:
- Individual convective cells using radar reflectivity data [Feng et al. (2022), MWR];
- MCSs using infrared brightness temperature (Tb) data from geostationary satellites, or outgoing longwave radiation (OLR) data from model simulations, with optional collocated precipitation data [Feng et al. (2021), JGR] or 3D radar reflectivity data [Feng et al. (2018) JAMES; Feng et al. (2019), JCLI] to identify robust MCSs;
- Generic 2D objects defined by customizable feature identification functions.
The input data must contain at least 3 dimensions: time, y, x, with corresponding coordinates of time, latitude, longitude. The latitude and longitude coordinates can be either 1D or 2D. But the data must be on a fixed 2D grid (any projection is fine) since PyFLEXTRKR only supports tracking data on 2D arrays. Irregular grids such as those in E3SM or MPAS model must first be regridded to a regular grid before tracking. Additional variable names and coordinate names are specified in the config file.
The dimension order in the input data does not need to be in time, y, x, as the dimensions are internally reordered when the data are read in.
- NEXRAD radar data
- ARM C-SAPR radar data
- GPM Tb+IMERG precipitation data
- WRF post-processed Tb + precipitation data
- E3SM regridded OLR + precipitation data
- ERA5 500hPa geopotential height anomaly data
An example Python script to map NEXRAD Level 2 data to a Cartesian grid netCDF file using PyART is provided in /pyflextrkr/grid_radar_pyart.py.
The gridded radar data produced by the example script can be used for convective cell tracking. An example of the data can be downloaded from: sample NEXRAD radar data.
Note that the terrain_file in the example radar cell tracking config file is optional. If terrain_file is provided, radar reflectivity data below surface_elevation + sfc_dz_min is filtered before calculating composite reflectivity to identify convective cells. This helps to minimize ground clutter and anomalous propagation effects on convective cell identification.
Generating the Terrain_Masking.nc netcdf file: Use the /pyflextrkr/make_terrain_rangemask.py script to generate the terrain masking file suited for the radar grids being used for cell tracking. Before running this script, you will need to download the topography (elevation data) file ETOPO1_Ice_g_gmt4.grd.gz from NOAA. Rename the file as ETOPO1_Ice_g_gmt4.nc after downloading and then run the python script to obtain the mask terrain output file.
An example run script for tracking MCSs directly from WRF output data is provided in the runscripts directory: /runscripts/run_mcs_tbpf_wrf.py.
The run script calls a pre-processing function for WRF data that produces Tb and rain rate for MCS tracking:
/pyflextrkr/preprocess_wrf_tb_rainrate.py
The pre-processing function works with standard WRF output data that contains OLR, RAINNC and RAINC. It converts OLR to Tb using a simple empirical relationship and calculates rain rates between consecutive times. An example config file for WRF MCS tracking is provide in /config/config_wrf4km_mcs_tbpf_example.yml.
For model simulation outputs that contains OLR and rain rate (unlike accumulated precipitation in WRF), set olr2tb : True to convert OLR [W/m^2] to Tb [K], and provide pcp_convert_factor to convert rain rate to the unit of [mm/hour] in the config file. See example config file: /config/config_model25km_mcs_tbpf_example.yml
For tracking generic features, a reader code is needed to produce the variables listed in Table 1.
Table 1. Variables required for generic feature tracking
| Variable Name in config file | Example Generic Name | Explanation |
|---|---|---|
| feature_varname | feature_mask | A 2D array with features of interest labeled by unique numbers. A simple example is labeling contiguous features with values larger than a threshold, using the SciPy function: scipy.ndimage.label. |
| nfeature_varname | nfeatures | Number of features in the file. |
| featuresize_varname | npix_feature | A 1D array with the number of pixels (i.e., size) for each labeled feature |
| time | Epoch time of the file |
An example of labeling generic features is provided in /pyflextrkr/idfeature_generic.py. The function contains two different methods for labeling features:
- Simple thresholds and connectivity (using ndimage.label function)
- Watershed segmentation (using skimage.watershed function)
After providing the reader code, add it to the idefeature_driver.py, and specify the feature_type in the config file (see example /config/config_era5_z500_example.yml). Here’s an example for generic feature identification:
if feature_type == "generic":
from pyflextrkr.idfeature_generic import idfeature_generic as id_featureWith this reader code, PyFLEXTRKR will run for any generic feature tracking and produce track statistics and labeled tracked numbers on the native grid (see Section 3 Algorithm and workflow and Figure 1). The track statistics contains basic statistics such as track_duration, base_time, meanlat, meanlon, area, etc. If more feature-specific statistics is desired, they can be added in /pyflextrkr/trackstats_func.py. All added track statistics variables in that function will be written in the output track statistics files automatically by the /pyflextrkr/trackstats_driver.py. Refer to the examples from feature_type == ‘tb_pf’ or ‘radar_cells’ in that function.
To run the code, type the following in the command line:
Activate PyFLEXTRKR virtual environment (see README.md on how to create a virtual environment and install PyFLEXTRKR):
conda activate flextrkrRun PyFLEXTRKR:
python run_mcs_tbpf.py config.ymlThe flags in Table 2 and Table 3 control each of the steps to be run, and they should be set to True to run the desired steps. For more detail explanations of the steps, refer to Section 3 Algorithm and workflow and Figure 1 and Figure 2.
Table 2. Controls for each tracking steps for all feature tracking.
| Parameter | Explanation |
|---|---|
| run_idfeature | Step 1: Identify features from input data |
| run_tracksingle | Step 2: Link features between consecutive pairs of times |
| run_gettracks | Step 3: Assign track numbers to linked features during the tracking period. |
| run_trackstats | Step 4: Calculate track statistics |
| run_mapfeature | Step 5: Map tracked feature numbers to native pixel files |
Table 3. Controls for each tracking steps for MCS tracking.
| Parameter | Explanation |
|---|---|
| run_idfeature | Step 1: Identify features from input data |
| run_tracksingle | Step 2: Link features between consecutive pairs of times |
| run_gettracks | Step 3: Assign track numbers to linked features during the tracking period |
| run_trackstats | Step 4: Calculate track statistics |
| run_identifymcs | Step 5: Identify MCS based on Tb data |
| run_matchpf | Step 6: Calculate PF statistics within tracked MCS |
| run_robustmcs | Step 7: Identify robust MCS based on PF characteristics |
| run_mapfeature | Step 8: Map tracked MCS numbers to native pixel files |
| run_speed | Step 9: Calculate MCS movement statistics |
The key parameters in the config file that need to be changed before running PyFLEXTRKR are listed in Table 4.
Table 4. Key parameters in the config file.
| Parameter | Explanation |
|---|---|
| startdate | Start date/time of tracking. E.g., '20200101.0000' |
| enddate | End date/time of tracking. E.g., '20200901.0000' |
| time_format | Time format of the input data file name. E.g., wrf_tb_rainrate_2020-01-01_00:00:00.nc time_format should be 'yyyy-mo-dd_hh:mm:ss' |
| databasename | String before the time string in the input data file name. E.g., wrf_tb_rainrate_2020-01-01_00:00:00.nc, databasename should be 'wrf_tb_rainrate_' |
| clouddata_path | Input data file directory |
| root_path | Tracking output files root directory. All files generated by the tracking will be written in this directory |
| pixel_radius | Spatial resolution of input data [km]. This is an approximated grid size and it is assumed to be the same across the entire domain |
| datatimeresolution | Temporal resolution of input data [hour] |
| landmask_filename | Land mask netCDF file name (optional). If provided, then tracked MCS statistics will have a pf_landfrac variable that can be used to distinguish MCS over land or ocean. Set this to an empty string “” if no land mask file is available |
| landmask_varname | Land mask variable name (optional) |
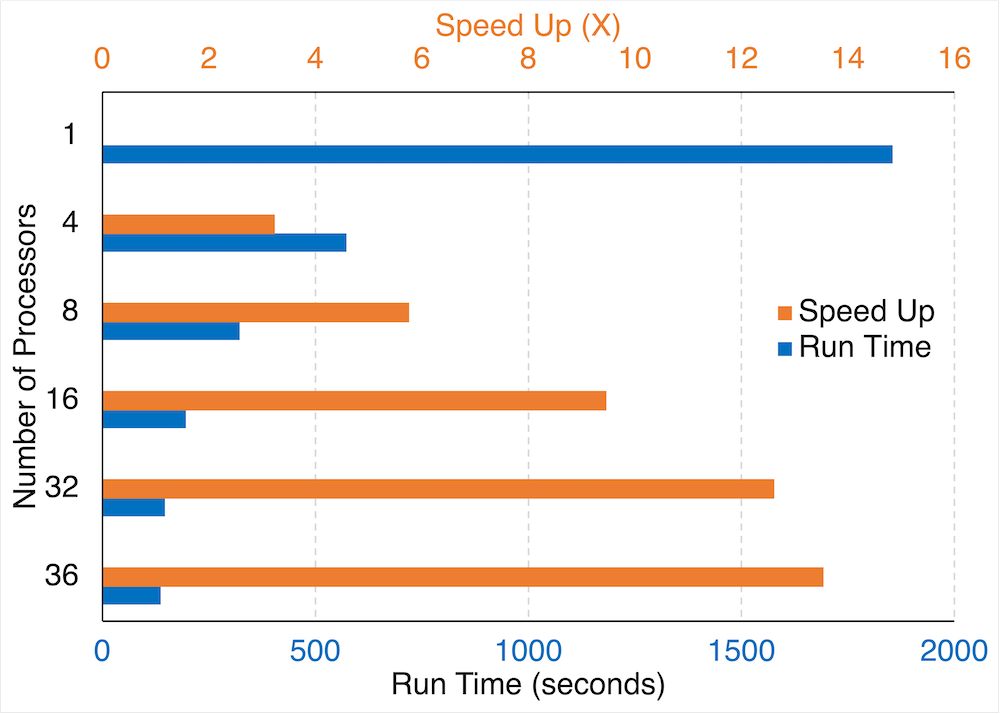
Running the code in parallel mode significantly reduces the time it takes to finish, particularly for larger datasets and/or longer continuous tracking period. For example, the figure below shows the performance scaling of tracking MCSs over South America for a one-month period using different number of processors (CPUs). Running with 16 processors (parallel) results in ~10x speed up compared to using a single processor (serial), cutting down the processing time from ~30 min (serial) to ~3 min (parallel). The performance scaling varies with the size of the dataset, but larger datasets likely scales better with more processors. The size of the dataset used in this performance test is moderate to small (690 x 480 pixels with 744 time frames).
There are two parallel options, controlled by setting the run_parallel value, as explained in Table 5.
Table 5. Parallel processing options.
| Parameter | Explanation |
|---|---|
| run_parallel | 0: run in serial. 1: use Dask LocalCluster (on multi-CPU computers, workstations) 2: use Dask distributed (on HPC clusters) |
| nprocesses | Number of processors to use. Only applicable if run_parallel=1. |
| timeout | Dask distributed timeout limit [second]. Only applicable if run_parallel=2. |
Note that running the code in parallel shares the total system memory available among the number of processors. For very large datasets such as global high resolution data (e.g., 3600x1800 pixels), this may result in out-ot-memory error if the number of tracks is too large (e.g., tracking for 1 year with hourly data). In that case, reducing the number of processors usually helps.
Running Dask distributed is an experimental feature and the capability is still being tested. Setting run_parallel=2 requires providing a Dask scheduler json file at run time like this:
python run_mcs_tbpf.py config.yml scheduler.jsonThe scheduler file can be created by:
srun -N 10 --ntasks-per-node=16 dask-worker
--scheduler-file=$SCRATCH/scheduler.json
--memory-limit='6GB'
--worker-class distributed.Worker
--local-directory=/tmp &Or by using dask-mpi:
srun -u dask-mpi \
--scheduler-file=$SCRATCH/scheduler.json
--nthreads=1
--memory-limit='auto'
--worker-class distributed.Worker
--local-directory=/tmp &Refer to the slurm script (under /slurm directory) to see an example set up on the DOE NERSC system.
Expected output files at the completion of generic feature tracking are listed in Table 6.
Table 6. Expected output files for generic feature tracking.
| Directory | File Names | Explanation |
|---|---|---|
stats_path_name (Track Statistics) |
tracknumbers_startdate_enddate.nc |
Track numbers output file from Step 3. |
stats_path_name (Track Statistics) |
trackstats_sparse_startdate_enddate.nc |
Track statistics output file from Step 4 (default sparse format). |
stats_path_name (Track Statistics) |
trackstats_startdate_enddate.nc |
Track statistics output file from Step 4 (optional dense format). |
pixel_path_name (Track mask pixel files) |
[pixeltracking_filebase]datetime.nc |
Individual pixel files containing track number masks from Step 5. |
To run tracking on climate data (e.g., multiple years), an example script is provided to demonstrate how to create multiple config files for a range of specified years and slurm job submission scripts.
The script replaces the STARTDATE and ENDDATE in a config template and a slurm template with a specific year, and saves them to new files for submitting as slurm jobs.
Similarly, an example script is provided to demonstrate how to post process large amount of tracking outputs to get monthly means. The example script creates a tasklist for calculating multiple years of monthly mean MCS statistics files that can be run in parallel using TaskFarmer on DOE's HPC system NERSC. An example slurm script using TaskFarmer to run the tasklist is provided here. The post processed monthly data can then be further analyzed and visualized, see Gallery of Statistical Analysis for more examples.
Tracking in PyFLEXTRKR primarily uses object overlap technique, with an option to use advection estimates (2D cross-correlation) to increase overlap probability. Largest overlap objects are tracked continuously, and smaller overlap objects are marked as merging/splitting.
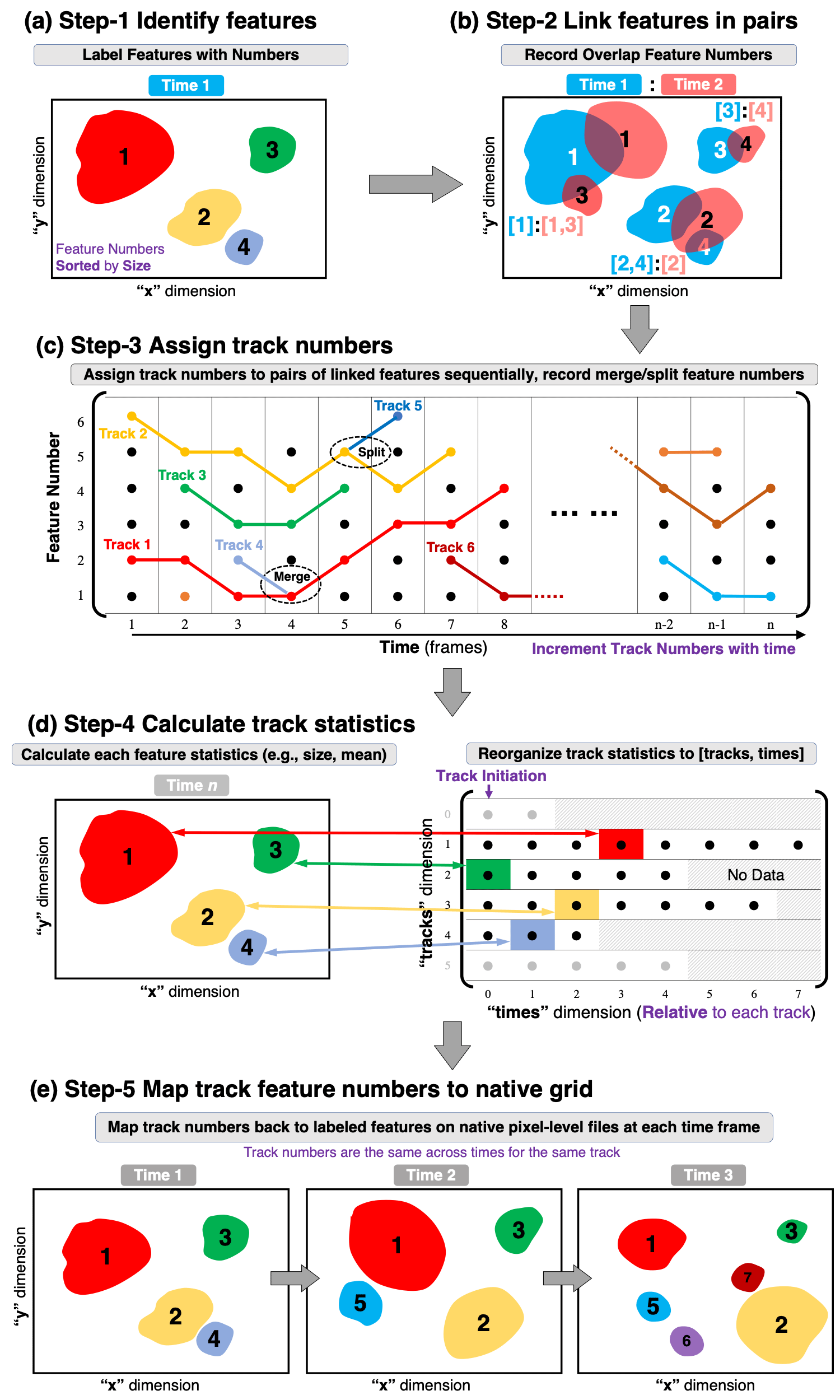
The main workflow of PyFLEXTRKR is illustrated in Figure 1. Explanation on the purpose for each of the steps are provided below.
Identify and label features of interest from individual time frames (Figure 1a).
Output: tracking_path_name/cloudid_yyyymmdd_hhmmss.nc
Link features between two consecutive time steps by checking their spatial overlap. If two features from consecutive timesteps (e.g., Feature #3 in Time 1 and feature #4 in Time 2) have an overlap fraction of more than X (othresh in config), they are connected in time and their numbers are recorded in pairs ([3]:[4]). If more than one feature at a time overlaps with a single feature at an adjacent time, they are all recorded (Figure 1b).
Output: tracking_path_name/track_yyyymmdd_hhmmss.nc
Extend the linked feature pairs between two consecutive time steps from Step 2 to the entire tracking period and assign track numbers. For example, these pairs of feature numbers are linked from time 1 through time 8: [2]:[2] (time 1-2), [2]:[1] (time 2-3), [1]:[1] (time 3-4), [1]:[2] (time 4-5), [2]:[3] (time 5-6), [3]:[3] (time 6-7), [3]:[4] (time 7-8), these features are assigned Track #1 (red color track in Figure 1c). Track numbers are incremented with time as each pair of consecutively linked features are processed. To consider situations when two or more features in one timestep are linked to the same feature in another timestep, the largest feature that overlaps is labeled as the continuation of the same track, and those smaller features are labeled as merging and/or splitting of the main track. For example, Track #4 merges with Track #1 at time 4 (light blue color track in Figure 1c), and Track 5 splits from Track #2 at time 5 (dark blue color track in Figure 1c).
Output: stats_path_name/tracknumbers_startdate_enddate.nc
Reorganize tracks to a format [tracks, times]. The “tracks” dimension contains the track number, and the “times” dimension is the relative time for each track. That is, times=0 is the initiation time for each track. Square dense arrays are created to store various statistics for the tracks, if a track duration is shorter than the “times” dimension, they are filled with missing values (hatched color showing “No Data” in Figure 1d).
For features at the same time, the feature identification file created in Step-1 is processed to calculate various statistics and put back to the [tracks, times] format (denoted by color arrows and color blocks in Figure 1d), such as location, size, etc.
In parallel processing, each feature identification file is handled by a task, after the statistics are collected when all the tasks are completed, a single netCDF file containing the track statistics is written. By default, a sparse array format netCDF is written for 2D variables (those that change by [tracks, times], e.g., base_time, area, etc.) to reduce memory usage and output file size. Optional dense (square) array format can be written by setting trackstats_dense_netcdf=1 in the config file. A function is also provided in ft_functions.py (convert_trackstats_sparse2dense) to convert sparse track statistics file to dense format.
Output: stats_path_name/trackstats_startdate_enddate.nc
Writes the track numbers back to the labeled feature masks on the native pixel-level files at each time. Each labeled feature from Step-1 is written with a unique track number during the tracking period, so that they are the same for the same track across different times (e.g., same color patches denote the same tracked feature in Figure 1e).
In parallel processing, the track numbers belonging to the same time are first read from the trackstats file from Step-4, then they are sent to a task to match the feature identification file from Step-1, and a netCDF file is written by the task.
Output: pixel_path_name/pixeltracking_filebase_yyyymmdd_hhmmss.nc
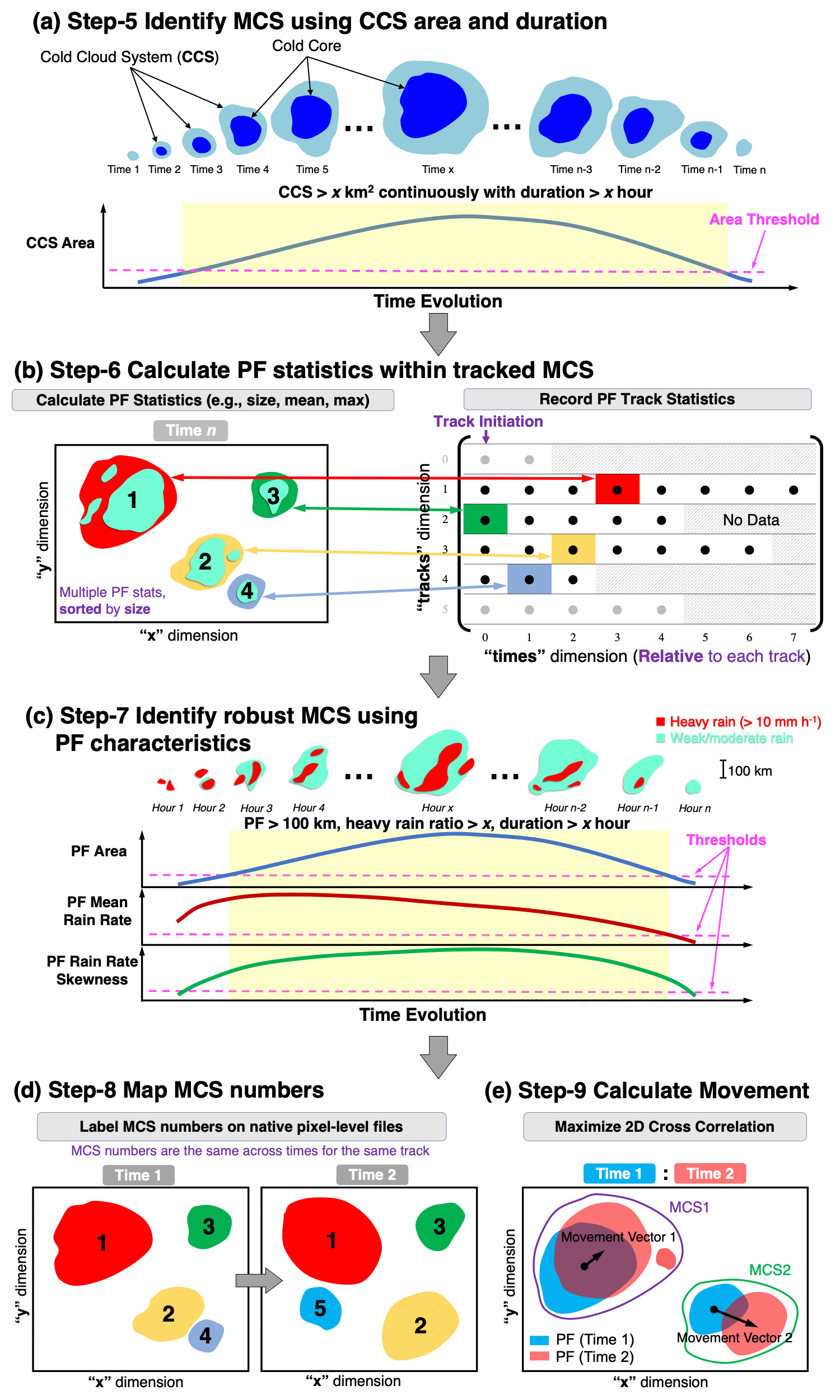
Tracking of MCS consists of a total of nine steps. The first four steps are the same as that shown in Figure 1, and the additional 5 steps are shown in Figure 2. Tracking is performed primarily on infrared brightness temperature (Tb) defined cold cloud systems (CCSs, which include cold cloud cores and cold anvils), with optional information provided by precipitation data to improve the identification of robust MCSs.
Since the first 4 steps are the same as tracking any features, the additional steps 5-9 specifically designed for MCSs are explained below:
Identify MCSs based on the CCS area and duration criteria. A track with CCS area > x km2 and persists for longer than x hour, and contains a cold core is defined as an MCS (Figure 2a). Tracks that meet MCS criteria are kept in the track statistics file. Smaller CCSs that merge with or split from those MCSs are also kept. Other tracks that are not associated with MCSs are removed. The CCS thresholds are set in the config file.
If there is no precipitation data available with the Tb data, this step is considered the final step of the MCS identification. Some modification of the code in Step 8 (see below) is needed to map the tracked MCS number to the pixel-level files.
Output: stats_outpath/mcs_tracks_startdate_enddate.nc
Match the collocated precipitation data within MCS cloud masks (including merges and splits) and calculate associated PF statistics, such as PF area, PF major axis length, mean rain rate, rain rate skewness, etc., and record to the track statistics file (Figure 2b). Providing an optional land mask input file in this step will yield PF land fraction in the output that can be used to separate land vs. ocean MCSs. In parallel processing, each cloudid file containing precipitation (produced in Step 1) is handled by a task, after all the PF statistics are collected after the tasks are completed, a single netCDF file containing the original CCS track statistics and the new PF statistics is written.
Output: stats_path_name/mcs_tracks_pf_startdate_enddate.nc
Identify robust MCSs based on the PF statistics and only keep the tracks that are robust MCSs. A track with PF major axis length > 100 km, with PF area, PF mean rain rate, PF rain rate skewness, and heavy rain ratio larger than lifetime dependent thresholds is defined as a robust MCS (Figure 2c). The PF thresholds are set in the config file.
Output: stats_path_name/mcs_tracks_robust_startdate_enddate.nc
Map the robust MCS track numbers back to original pixel-level domain at each time step (Figure 2d). The original pixel-level IR and precipitation data are also stored in the output.
Output: pixel_path_name/startdate_enddate/mcstrack_yyyymmdd_hhmmss.nc
Calculate robust MCS movement statistics such as movement speed, direction, and add it to the MCS track statistics file (Figure 2e).
Output: stats_path_name/mcs_tracks_final_startdate_enddate.nc