diff --git a/FAQ.md b/FAQ.md
new file mode 100644
index 0000000..f9fc56e
--- /dev/null
+++ b/FAQ.md
@@ -0,0 +1,73 @@
+# Can we use the SAS university edition for workshops? #
+
+We are looking into that. But probably!
+
+# I have a Mac, can I use SAS? #
+
+With the SAS University Edition you can. We personally have not tried
+it, but it does say it can work for Mac's.
+
+# How do I install SAS on a laptop without a CD drive? #
+
+If you bought the SAS CD, it comes with a License key that you can use
+to download SAS using the SAS Download Manager.
+
+# Message `sh.exe: nano: command not found`. Help? #
+
+> I was just practicing what we’ve done last Tuesday and I am getting
+> this message for nano command: sh.exe”: nano: command not found. It
+> worked well in class, but is not working now. Would you happen to know
+> the solution? I did specify at the beginning git config --global
+> core.editor "notepad"
+
+Thanks for your question. Looks like your git was configured to use
+Notepad as the text editor, not Nano. Try replacing "nano" with
+"notepad" in your code and see if that works.
+
+# What is a repository? #
+
+> What exactly a repository is? Is it that folder we created named
+> “practice”?
+
+A repository is a history of all your commits. Everytime you do `git
+commit`, it saves the files you are committing into the
+repository. The physical repository is the `.git` folder, but it
+tracks everything in that folder (the "practice" folder).
+
+# Why do we need to check the repo's status so often? #
+
+Checking often is important to make sure you are aware of what is
+going on in your repository.
+
+# What exactly the "untracked" means? #
+
+"Untracked" means that the file is seen by git, but is not
+followed. So any changes to the file will not be recognized by git
+*until* you follow it (track it). (Like following people on Facebook
+or twitter. They exist when you aren't following them, but you don't
+keep updated on their status or what they are doing.).
+
+# What happens with the saved files but not committed? #
+
+Saved files (`git add` you mean?) are put into the staging area so
+that they can be committed into the history (the git repository). If
+you don't commit them, they stay there, but they do *not* go into the
+history. Modified files (when you *don't* `git add` them, continue to
+stay modified as you edit them, but they do not get saved until you do
+`git add` and `git commit`.
+
+# 'No new line at the end of file' after running `git diff` #
+
+This can easily be fixed by pressing Enter at the bottom of the file.
+This occurs because (more or less) Git follows the Unix convention.
+If you want a more detailed answer, see
+[this answer on the why](http://stackoverflow.com/a/5813359/2632184)
+and
+[this answer on the history of why](http://stackoverflow.com/questions/729692/why-should-files-end-with-a-newline).
+
+# My terminal is stuck and I can't get back to the prompt (`$` or `>`) #
+
+If you ever get stuck in the terminal, hit `Control-c` to 'c'ancel the
+command and get back to the prompt (the `$` or `>` characters).
+
+
diff --git a/LICENSE.md b/LICENSE.md
new file mode 100644
index 0000000..7ff305e
--- /dev/null
+++ b/LICENSE.md
@@ -0,0 +1,183 @@
+---
+layout: default
+title: License
+---
+
+> Content: Copyright (c) 2015 Luke W. Johnston, Daiva Nielsen, and Sarah Meister (CC-BY)
+>
+> Design: Copyright (c) 2013 Mark Otto (MIT, see bottom)
+
+Copyright (c) 2015 Luke W. Johnston and Daiva Nielsen (CC-BY)
+
+## creative commons
+
+# Attribution 4.0 International
+
+Creative Commons Corporation (“Creative Commons”) is not a law firm and does not provide legal services or legal advice. Distribution of Creative Commons public licenses does not create a lawyer-client or other relationship. Creative Commons makes its licenses and related information available on an “as-is” basis. Creative Commons gives no warranties regarding its licenses, any material licensed under their terms and conditions, or any related information. Creative Commons disclaims all liability for damages resulting from their use to the fullest extent possible.
+
+### Using Creative Commons Public Licenses
+
+Creative Commons public licenses provide a standard set of terms and conditions that creators and other rights holders may use to share original works of authorship and other material subject to copyright and certain other rights specified in the public license below. The following considerations are for informational purposes only, are not exhaustive, and do not form part of our licenses.
+
+* __Considerations for licensors:__ Our public licenses are intended for use by those authorized to give the public permission to use material in ways otherwise restricted by copyright and certain other rights. Our licenses are irrevocable. Licensors should read and understand the terms and conditions of the license they choose before applying it. Licensors should also secure all rights necessary before applying our licenses so that the public can reuse the material as expected. Licensors should clearly mark any material not subject to the license. This includes other CC-licensed material, or material used under an exception or limitation to copyright. [More considerations for licensors](http://wiki.creativecommons.org/Considerations_for_licensors_and_licensees#Considerations_for_licensors).
+
+* __Considerations for the public:__ By using one of our public licenses, a licensor grants the public permission to use the licensed material under specified terms and conditions. If the licensor’s permission is not necessary for any reason–for example, because of any applicable exception or limitation to copyright–then that use is not regulated by the license. Our licenses grant only permissions under copyright and certain other rights that a licensor has authority to grant. Use of the licensed material may still be restricted for other reasons, including because others have copyright or other rights in the material. A licensor may make special requests, such as asking that all changes be marked or described. Although not required by our licenses, you are encouraged to respect those requests where reasonable. [More considerations for the public](http://wiki.creativecommons.org/Considerations_for_licensors_and_licensees#Considerations_for_licensees).
+
+## Creative Commons Attribution 4.0 International Public License
+
+By exercising the Licensed Rights (defined below), You accept and agree to be bound by the terms and conditions of this Creative Commons Attribution 4.0 International Public License ("Public License"). To the extent this Public License may be interpreted as a contract, You are granted the Licensed Rights in consideration of Your acceptance of these terms and conditions, and the Licensor grants You such rights in consideration of benefits the Licensor receives from making the Licensed Material available under these terms and conditions.
+
+### Section 1 – Definitions.
+
+a. __Adapted Material__ means material subject to Copyright and Similar Rights that is derived from or based upon the Licensed Material and in which the Licensed Material is translated, altered, arranged, transformed, or otherwise modified in a manner requiring permission under the Copyright and Similar Rights held by the Licensor. For purposes of this Public License, where the Licensed Material is a musical work, performance, or sound recording, Adapted Material is always produced where the Licensed Material is synched in timed relation with a moving image.
+
+b. __Adapter's License__ means the license You apply to Your Copyright and Similar Rights in Your contributions to Adapted Material in accordance with the terms and conditions of this Public License.
+
+c. __Copyright and Similar Rights__ means copyright and/or similar rights closely related to copyright including, without limitation, performance, broadcast, sound recording, and Sui Generis Database Rights, without regard to how the rights are labeled or categorized. For purposes of this Public License, the rights specified in Section 2(b)(1)-(2) are not Copyright and Similar Rights.
+
+d. __Effective Technological Measures__ means those measures that, in the absence of proper authority, may not be circumvented under laws fulfilling obligations under Article 11 of the WIPO Copyright Treaty adopted on December 20, 1996, and/or similar international agreements.
+
+e. __Exceptions and Limitations__ means fair use, fair dealing, and/or any other exception or limitation to Copyright and Similar Rights that applies to Your use of the Licensed Material.
+
+f. __Licensed Material__ means the artistic or literary work, database, or other material to which the Licensor applied this Public License.
+
+g. __Licensed Rights__ means the rights granted to You subject to the terms and conditions of this Public License, which are limited to all Copyright and Similar Rights that apply to Your use of the Licensed Material and that the Licensor has authority to license.
+
+h. __Licensor__ means the individual(s) or entity(ies) granting rights under this Public License.
+
+i. __Share__ means to provide material to the public by any means or process that requires permission under the Licensed Rights, such as reproduction, public display, public performance, distribution, dissemination, communication, or importation, and to make material available to the public including in ways that members of the public may access the material from a place and at a time individually chosen by them.
+
+j. __Sui Generis Database Rights__ means rights other than copyright resulting from Directive 96/9/EC of the European Parliament and of the Council of 11 March 1996 on the legal protection of databases, as amended and/or succeeded, as well as other essentially equivalent rights anywhere in the world.
+
+k. __You__ means the individual or entity exercising the Licensed Rights under this Public License. Your has a corresponding meaning.
+
+### Section 2 – Scope.
+
+a. ___License grant.___
+
+ 1. Subject to the terms and conditions of this Public License, the Licensor hereby grants You a worldwide, royalty-free, non-sublicensable, non-exclusive, irrevocable license to exercise the Licensed Rights in the Licensed Material to:
+
+ A. reproduce and Share the Licensed Material, in whole or in part; and
+
+ B. produce, reproduce, and Share Adapted Material.
+
+ 2. __Exceptions and Limitations.__ For the avoidance of doubt, where Exceptions and Limitations apply to Your use, this Public License does not apply, and You do not need to comply with its terms and conditions.
+
+ 3. __Term.__ The term of this Public License is specified in Section 6(a).
+
+ 4. __Media and formats; technical modifications allowed.__ The Licensor authorizes You to exercise the Licensed Rights in all media and formats whether now known or hereafter created, and to make technical modifications necessary to do so. The Licensor waives and/or agrees not to assert any right or authority to forbid You from making technical modifications necessary to exercise the Licensed Rights, including technical modifications necessary to circumvent Effective Technological Measures. For purposes of this Public License, simply making modifications authorized by this Section 2(a)(4) never produces Adapted Material.
+
+ 5. __Downstream recipients.__
+
+ A. __Offer from the Licensor – Licensed Material.__ Every recipient of the Licensed Material automatically receives an offer from the Licensor to exercise the Licensed Rights under the terms and conditions of this Public License.
+
+ B. __No downstream restrictions.__ You may not offer or impose any additional or different terms or conditions on, or apply any Effective Technological Measures to, the Licensed Material if doing so restricts exercise of the Licensed Rights by any recipient of the Licensed Material.
+
+ 6. __No endorsement.__ Nothing in this Public License constitutes or may be construed as permission to assert or imply that You are, or that Your use of the Licensed Material is, connected with, or sponsored, endorsed, or granted official status by, the Licensor or others designated to receive attribution as provided in Section 3(a)(1)(A)(i).
+
+b. ___Other rights.___
+
+ 1. Moral rights, such as the right of integrity, are not licensed under this Public License, nor are publicity, privacy, and/or other similar personality rights; however, to the extent possible, the Licensor waives and/or agrees not to assert any such rights held by the Licensor to the limited extent necessary to allow You to exercise the Licensed Rights, but not otherwise.
+
+ 2. Patent and trademark rights are not licensed under this Public License.
+
+ 3. To the extent possible, the Licensor waives any right to collect royalties from You for the exercise of the Licensed Rights, whether directly or through a collecting society under any voluntary or waivable statutory or compulsory licensing scheme. In all other cases the Licensor expressly reserves any right to collect such royalties.
+
+### Section 3 – License Conditions.
+
+Your exercise of the Licensed Rights is expressly made subject to the following conditions.
+
+a. ___Attribution.___
+
+ 1. If You Share the Licensed Material (including in modified form), You must:
+
+ A. retain the following if it is supplied by the Licensor with the Licensed Material:
+
+ i. identification of the creator(s) of the Licensed Material and any others designated to receive attribution, in any reasonable manner requested by the Licensor (including by pseudonym if designated);
+
+ ii. a copyright notice;
+
+ iii. a notice that refers to this Public License;
+
+ iv. a notice that refers to the disclaimer of warranties;
+
+ v. a URI or hyperlink to the Licensed Material to the extent reasonably practicable;
+
+ B. indicate if You modified the Licensed Material and retain an indication of any previous modifications; and
+
+ C. indicate the Licensed Material is licensed under this Public License, and include the text of, or the URI or hyperlink to, this Public License.
+
+ 2. You may satisfy the conditions in Section 3(a)(1) in any reasonable manner based on the medium, means, and context in which You Share the Licensed Material. For example, it may be reasonable to satisfy the conditions by providing a URI or hyperlink to a resource that includes the required information.
+
+ 3. If requested by the Licensor, You must remove any of the information required by Section 3(a)(1)(A) to the extent reasonably practicable.
+
+ 4. If You Share Adapted Material You produce, the Adapter's License You apply must not prevent recipients of the Adapted Material from complying with this Public License.
+
+### Section 4 – Sui Generis Database Rights.
+
+Where the Licensed Rights include Sui Generis Database Rights that apply to Your use of the Licensed Material:
+
+a. for the avoidance of doubt, Section 2(a)(1) grants You the right to extract, reuse, reproduce, and Share all or a substantial portion of the contents of the database;
+
+b. if You include all or a substantial portion of the database contents in a database in which You have Sui Generis Database Rights, then the database in which You have Sui Generis Database Rights (but not its individual contents) is Adapted Material; and
+
+c. You must comply with the conditions in Section 3(a) if You Share all or a substantial portion of the contents of the database.
+
+For the avoidance of doubt, this Section 4 supplements and does not replace Your obligations under this Public License where the Licensed Rights include other Copyright and Similar Rights.
+
+### Section 5 – Disclaimer of Warranties and Limitation of Liability.
+
+a. __Unless otherwise separately undertaken by the Licensor, to the extent possible, the Licensor offers the Licensed Material as-is and as-available, and makes no representations or warranties of any kind concerning the Licensed Material, whether express, implied, statutory, or other. This includes, without limitation, warranties of title, merchantability, fitness for a particular purpose, non-infringement, absence of latent or other defects, accuracy, or the presence or absence of errors, whether or not known or discoverable. Where disclaimers of warranties are not allowed in full or in part, this disclaimer may not apply to You.__
+
+b. __To the extent possible, in no event will the Licensor be liable to You on any legal theory (including, without limitation, negligence) or otherwise for any direct, special, indirect, incidental, consequential, punitive, exemplary, or other losses, costs, expenses, or damages arising out of this Public License or use of the Licensed Material, even if the Licensor has been advised of the possibility of such losses, costs, expenses, or damages. Where a limitation of liability is not allowed in full or in part, this limitation may not apply to You.__
+
+c. The disclaimer of warranties and limitation of liability provided above shall be interpreted in a manner that, to the extent possible, most closely approximates an absolute disclaimer and waiver of all liability.
+
+### Section 6 – Term and Termination.
+
+a. This Public License applies for the term of the Copyright and Similar Rights licensed here. However, if You fail to comply with this Public License, then Your rights under this Public License terminate automatically.
+
+b. Where Your right to use the Licensed Material has terminated under Section 6(a), it reinstates:
+
+ 1. automatically as of the date the violation is cured, provided it is cured within 30 days of Your discovery of the violation; or
+
+ 2. upon express reinstatement by the Licensor.
+
+ For the avoidance of doubt, this Section 6(b) does not affect any right the Licensor may have to seek remedies for Your violations of this Public License.
+
+c. For the avoidance of doubt, the Licensor may also offer the Licensed Material under separate terms or conditions or stop distributing the Licensed Material at any time; however, doing so will not terminate this Public License.
+
+d. Sections 1, 5, 6, 7, and 8 survive termination of this Public License.
+
+### Section 7 – Other Terms and Conditions.
+
+a. The Licensor shall not be bound by any additional or different terms or conditions communicated by You unless expressly agreed.
+
+b. Any arrangements, understandings, or agreements regarding the Licensed Material not stated herein are separate from and independent of the terms and conditions of this Public License.
+
+### Section 8 – Interpretation.
+
+a. For the avoidance of doubt, this Public License does not, and shall not be interpreted to, reduce, limit, restrict, or impose conditions on any use of the Licensed Material that could lawfully be made without permission under this Public License.
+
+b. To the extent possible, if any provision of this Public License is deemed unenforceable, it shall be automatically reformed to the minimum extent necessary to make it enforceable. If the provision cannot be reformed, it shall be severed from this Public License without affecting the enforceability of the remaining terms and conditions.
+
+c. No term or condition of this Public License will be waived and no failure to comply consented to unless expressly agreed to by the Licensor.
+
+d. Nothing in this Public License constitutes or may be interpreted as a limitation upon, or waiver of, any privileges and immunities that apply to the Licensor or You, including from the legal processes of any jurisdiction or authority.
+
+> Creative Commons is not a party to its public licenses. Notwithstanding, Creative Commons may elect to apply one of its public licenses to material it publishes and in those instances will be considered the “Licensor.” Except for the limited purpose of indicating that material is shared under a Creative Commons public license or as otherwise permitted by the Creative Commons policies published at [creativecommons.org/policies](http://creativecommons.org/policies), Creative Commons does not authorize the use of the trademark “Creative Commons” or any other trademark or logo of Creative Commons without its prior written consent including, without limitation, in connection with any unauthorized modifications to any of its public licenses or any other arrangements, understandings, or agreements concerning use of licensed material. For the avoidance of doubt, this paragraph does not form part of the public licenses.
+
+> Creative Commons may be contacted at [creativecommons.org](http://creativecommons.org/).
+
+<<<<<<< HEAD
+# Released under MIT License for Poole
+
+Copyright (c) 2013 Mark Otto for website layout and design.
+
+Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the "Software"), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions:
+
+The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software.
+
+THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.
+=======
+>>>>>>> master
diff --git a/README.md b/README.md
new file mode 100644
index 0000000..30da74d
--- /dev/null
+++ b/README.md
@@ -0,0 +1,26 @@
+Code As Manuscript Workshops
+============================
+
+This series of *hands-on* workshops aims to introduce students to the
+concept of research reproducibility and to get the students practicing
+with using version control systems and with using modern techniques in
+R (or SAS) that make analyses less error prone and reproducible, and
+that make you more efficient, productive, and in control. Techniques
+for using version control systems to faciliate greater collaboration
+among peers are also taught. The ultimate goal of the workshop is to
+show how to reduce the number of steps needed to go from the initial
+data analysis to the final written manuscript or thesis (hence the
+name *Code As Manuscript*).
+
+The `workshops` repository is where the material is developed and
+sent to the website (which is on the `gh-pages` branch).
+
+To contribute, fork the repository and submit a pull request!
+
+# Resources for developing the lessons: #
+
+For images, put them in the `/lessons/images` folder and reference
+them within the lesson using `../images/`.
+
+For developing lesson plans, using the pull request feature. For
+maintaining the site and the repository, use direct pushing.
diff --git a/about.md b/about.md
new file mode 100644
index 0000000..9c15ee2
--- /dev/null
+++ b/about.md
@@ -0,0 +1,60 @@
+---
+layout: page
+title: About
+sidebar: true
+permalink: /
+---
+
+
+
+ Code As Manuscript is a group that teaches a series of workshops on
+ rethinking how code and analyses are structured, and how to
+ implement it. Our mission is to provide training to researchers and
+ scientists on how to make their analyses more reproducible and to
+ change how researchers view their code.
+
+
+
+We get much inspiration for our workshops from
+[Software Carpentry (SWC)](http://software-carpentry.org/), of which
+we are also SWC instructors.
+
+Currently, we teach these workshops to graduate students in our home
+department and are in the process of expanding to the
+[Graduate Professional Skills program](http://www.sgs.utoronto.ca/currentstudents/Pages/Professional-Development.aspx)
+at the University of Toronto. Our material is in general split into
+two parts:
+
+* [Lesson content](lessons/)
+* [GPS content](gps/), which also encompasses the lesson content
+
+## Misc facts about this website ##
+
+* Uses the [Hyde](http://hyde.getpoole.com/)
+ [Jekyll](http://jekyllrb.com) theme
+* Developed on GitHub and hosted for free on [GitHub Pages](https://pages.github.com)
+* The content on our site is [licensed](LICENSE/) under CC-BY, while
+ the website design is under MIT. Check out our
+ [license page](LICENSE/) for more info.
+
+Have questions or suggestions? Feel free to
+[open an issue on GitHub](https://github.com/codeasmanuscript/development/issues/new)
+or email the *Code As Manuscript* instructors () for
+more information
+
+# Site listing and content: #
+
+{% for cat in site.category-list %}
+
+## {{ cat }} ##
+
+
+ {% for page in site.pages %}
+ {% for pc in page.categories %}
+ {% if pc == cat %}
+ - {{ page.title }}
+ {% endif %}
+ {% endfor %}
+ {% endfor %}
+
+{% endfor %}
diff --git a/gps/about.md b/gps/about.md
new file mode 100644
index 0000000..443d70e
--- /dev/null
+++ b/gps/about.md
@@ -0,0 +1,20 @@
+---
+layout: page
+sidebar: true
+title: GPS
+permalink: gps/
+---
+
+GPS, or
+[Graduate Professional Skills program](http://www.sgs.utoronto.ca/currentstudents/Pages/Professional-Development.aspx),
+is offered by the
+[School of Graduate Studies](http://www.sgs.utoronto.ca/Pages/default.aspx)
+at the University of Toronto. The goal of the program is to provide
+workshops that teach students skills that may be useful in careers
+after they graduate. Completing the GPS program is recorded on the
+students' transcript. Our GPS-approved *Code As Manuscript* series of
+workshops provide credits to completion of the GPS program. Our links
+for the workshop series are:
+
+* [Syllabus](sas-syllabus/) for our SAS workshops.
+* [Syllabus](r-syllabus/) for our R workshops.
diff --git a/gps/r-syllabus.md b/gps/r-syllabus.md
new file mode 100644
index 0000000..71067c9
--- /dev/null
+++ b/gps/r-syllabus.md
@@ -0,0 +1,102 @@
+---
+title: "Code as Manuscript: Data wrangling, visualization, and reproducibility in R"
+author:
+ - Luke W. Johnston
+ - Sarah Meister
+date: 2015-10-17
+layout: page
+sidebar: false
+tag:
+ - GPS
+ - R
+ - Syllabus
+categories:
+ - GPS
+ - R
+ - Syllabus
+---
+
+## Course description: ##
+
+This series of *hands-on* workshops aims to introduce students to the
+concept of research reproducibility and to get the students practicing
+with using version control systems and with using modern techniques in
+R that make analyses less error prone and reproducible, and that make
+you more efficient, productive, and in control. Techniques for using
+version control systems to faciliate greater collaboration among peers
+will also be presented. The ultimate goal of the workshop is to show
+how to reduce the number of steps needed to go from the initial data
+analysis to the final written manuscript or thesis (hence the name
+*Code As Manuscript*). Given the applied nature of the concepts in
+these workshops, hands-on activities and
+[live coding](http://en.wikipedia.org/wiki/Live_coding) will be
+integrated into each workshop.
+
+## Goal: ##
+
+The expected goal of the workshops is that you will be able to:
+
+* Put your research under version control using Git.
+* Push and pull your git research repository to either
+ [GitHub](https://github.com/) or
+ [BitBucket](https://bitbucket.org/).
+* Learn the basics of R and functions within R
+* Produce publication quality plots
+* Quickly wrangle your data into an analyzable format
+* Reproducibly incorporate R code into your manuscript or thesis to
+ instantly add results and plots (no more copy and paste)
+
+Attaining these goals will be the first steps in making an efficient
+and highly productive workflow, that is also scientifically rigorous
+and transparent, and which you can take with you for the rest of your
+career (since R is free!).
+
+## Schedule ##
+
+Lesson content can be found [here]({{ site.github.url }}/lessons/). The workshop will
+follow the following sequence of topics on **Mondays from 3:00-6:00pm** at **65
+St. George St., room 201**:
+
+1. Git & GitHub -- Oct. 26th
+2. Basics of R -- Nov. 2nd
+3. Visualization -- Nov. 9th
+4. Data wrangling -- Nov. 16th
+5. Dynamic report generation -- Nov. 23rd
+
+## Intended audience: ##
+
+Graduate students or post-docs whose research involves a fair amount
+of data analysis. No experience necessary for these workshops.
+
+## Pre-requisites: ##
+
+* Fully charged laptop (though we do have outlets)
+* Install the appropriate software (see our
+ [instructions page, under the R section]({{ site.github.url }}/lessons/instructions/))
+* Bring a positive, not-afraid-of-making-mistakes-or-feeling-unsure
+ attitude!! Learning any language (either human or computer) is
+ hard work and *not* easy, but can be done!
+
+## Assignments: ##
+
+Because this is a hands-on workshop, at the end of each workshop, we
+have an activity for you to try out. And since this is a GPS-approved
+course, if you want to get a GPS credit, you will need to:
+
+1. Come to all the workshops (though we are flexible)
+2. Complete each workshop assignment and push to the shared repository
+ on GitHub (so we can track and view it).
+
+Don't worry if this doesn't make sense yet. We will go over all these
+details over the workshop series.
+
+# Instructors #
+
+Luke Johnston, MSc, PhD (c)
+luke.johnston@mail.utoronto.ca
+
+Sarah Meister, MSc (c)
+sarah.meister@mail.utoronto.ca
+
+You can contact the workshop email at: codeasmanuscript@gmail.com
+
diff --git a/gps/sas-syllabus.md b/gps/sas-syllabus.md
new file mode 100644
index 0000000..d9141b6
--- /dev/null
+++ b/gps/sas-syllabus.md
@@ -0,0 +1,109 @@
+---
+title: "Code as Manuscript: Practices for Reproducibility and Efficiency in SAS"
+author:
+ - Luke Johnston
+ - Daiva Nielsen
+date: 2015-06-15
+geometry: margin=1in
+fontsize: 12pt
+papersize: letterpaper
+layout: page
+sidebar: false
+tag:
+ - GPS
+ - SAS
+ - Syllabus
+categories:
+ - GPS
+ - SAS
+ - Syllabus
+---
+
+## Course description: ##
+
+This series of workshops aims to introduce students to the concept of
+research reproducibility and to get the students practicing with using
+version control systems and with using techniques in SAS that make the
+analysis more efficient, less error prone, and less stressful.
+Techniques for using version control systems to faciliate greater
+collaboration among peers will also be presented. The ultimate goal
+of the workshops is to demonstrate how to reduce the total number of
+steps required to produce a manuscript from the data analysis stage
+(hence the name *Code As Manuscript*). Given the applied nature of
+the concepts in these workshops, hands-on activities and
+[live coding](http://en.wikipedia.org/wiki/Live_coding) will be
+integrated into each workshop.
+
+## Goal: ##
+
+The expected goal of the workshops is that you will be able to:
+
+* Put your research under version control using Git.
+* Push and pull your git research repository to either
+ [GitHub](https://github.com/) or
+ [BitBucket](https://bitbucket.org/).
+* Simplify your SAS code by using macros and ODS facilities in a way
+ that allows code to output results into a file that can be easily
+ incorporated into a manuscript or report.
+* To put the macros into either a macro file or as a SAS autocall
+ library so that the macros are useable in all your SAS files.
+
+Attaining these goals will the first steps in making an efficient and
+highly productive workflow, that is also scientifically rigorous and
+transparent, which you can take with you for the rest of your career.
+
+## Schedule ##
+
+Lesson content can be found [here]({{ site.github.url }}/lessons/). The workshop will
+follow the following sequence of topics:
+
+1. Git: June 16th, 3:00-5:00pm
+2. GitHub: June 23rd, 3:00-5:00pm
+3. Macros: June 30th, 3:00-5:00pm
+4. ODS: July 7th, 3:00-5:00pm
+
+## Intended audience: ##
+
+Graduate students or post-docs whose research involves a fair amount
+of data analysis using SAS.
+
+## Pre-requisites: ##
+
+* Bring a fully charged laptop
+* Install the appropriate software (see our
+ [instructions page]({{ site.github.url }}/lessons/instructions/))
+* Fairly competent in coding SAS commands. *We are not here to teach
+ an introduction to SAS*, we assume you are fairly competent with
+ using SAS. If you are a novice, recognize that you may not
+ understand everything that is going on... *However*, you will still
+ likely learn a lot!!
+* Some familiarity with the command-line or with computing is
+ desirable, but not necessary.
+
+## Assignment: ##
+
+Because of the hands-on nature of the workshops, in order to obtain a
+GPS credit for the workshops you need to:
+
+1. Come to all the workshops
+2. Complete each workshop assignment
+
+To hand in the assignment, you will need to:
+
+1. Fork our shared workshop repository
+2. Clone it to your computer
+3. Create a new folder with your name
+4. Put the assignments for each workshop in your new folder
+5. Commit and push the assignments to your fork
+6. Submit a pull request to the main shared repository
+
+Don't worry if this doesn't make sense yet. We will go over all these
+details over the workshop series.
+
+# Instructors #
+
+Luke Johnston, MSc, PhD (c)
+luke.johnston@mail.utoronto.ca
+
+Daiva Nielsen, PhD
+daiva.nielsen@mail.utoronto.ca
diff --git a/lessons/assumptions/assignment.md b/lessons/assumptions/assignment.md
new file mode 100644
index 0000000..3d33a67
--- /dev/null
+++ b/lessons/assumptions/assignment.md
@@ -0,0 +1,21 @@
+---
+title: "Assignment: Statistical Assumptions"
+author:
+ - Luke
+ - Daiva
+date: 2015-03-20
+fontsize: 12pt
+geometry: margin=1in
+papersize: letterpaper
+layout: default
+tag:
+ - Lessons
+ - Assignment
+ - Assumptions
+categories:
+ - Lessons
+ - Assumptions
+
+---
+
+> Work in progress
diff --git a/lessons/assumptions/cheatsheet.md b/lessons/assumptions/cheatsheet.md
new file mode 100644
index 0000000..f33733a
--- /dev/null
+++ b/lessons/assumptions/cheatsheet.md
@@ -0,0 +1,84 @@
+## Cheatsheet: Statistical Assumptions ##
+
+This cheatsheet provides statistical codes that can be used in SAS to test 3 assumptions of linear regression: model fit, residual distribution and residual variance. It also provides statistical codes to examine 2 important factors to examine in statistical models: collinearity and influence/outliers (however, these are not assumptions of linear regression).
+
+## SAS codes for Assumptions##
+
+*To view contents of the SAS practice dataset "Class"*
+
+```
+proc contents data=sashelp.class;
+run;
+```
+
+*Assumption 1) Model Fit -- make a scatterplot*
+
+```
+proc sgplot data=sashelp.class;
+scatter x=height y=weight;
+run;
+```
+
+*Assumption 2) Residual Distribution (2 step process)*
+
+*Step 1: Run a linear regression and output residual and predicted terms in a new dataset*
+
+```
+proc reg data=sashelp.class;
+model height=weight age;
+output out=resid residual=r predicted=fit;
+run;
+quit;
+```
+
+*Step 2: Plot the output of the new dataset*
+
+```
+goptions reset=all;
+proc univariate data=resid normal;
+var r;
+qqplot r / normal(mu=est sigma=est);
+run;
+```
+
+*Assumption 3) Residual Variance -- run a linear regression and create a predicted plot*
+
+```
+proc reg data=sashelp.class;
+model height=weight / spec;
+plot r.*p.;
+run;
+quit;
+```
+
+## SAS codes for Collinearity and Outliers ##
+
+*Collinearity -- examine VIF or tolerance*
+
+```
+proc reg data=sashelp.class;
+model height = weight age / vif tol;
+run;
+quit;
+```
+
+*Outliers -- make a scatterplot and visually inspect the points or run proc univariate to obtain 5 highest and 5 lowest extreme observations*
+
+*Scatterplot -- this is a more complex code to make a scatterplot. Using the code from "Assumption 1) Model Fit" will also work.*
+
+```
+proc gplot data=sashelp.class;
+plot height*weight=1 / vaxis=axis1;
+run;
+quit;
+```
+
+*Proc univariate to obtain output of extreme observations*
+
+```
+proc univariate data=sashelp.class;
+var height weight;
+run;
+quit;
+```
+
diff --git a/lessons/assumptions/intro.md b/lessons/assumptions/intro.md
new file mode 100644
index 0000000..9d987a7
--- /dev/null
+++ b/lessons/assumptions/intro.md
@@ -0,0 +1,8 @@
+# Outline of workshop #
+
+* Purpose: Familiarize students with some assumptions of linear regression & teach how they can check them using SAS.
+* Specifically we cover these 3 assumptions: model fit, residual distribution and residual variance.
+* Provide SAS codes that are used to examine data to see if assumptions are satisfied.
+* Do not go into depth on 2 other assumptions (independence & error), but we explain what these mean and direct students to other resources about these assumptions.
+* Also cover collinearity and influence/outliers. These are not assumptions, but important factors to examine in statistical models.
+* Workshop exercises (live-coding): 1) Workshop instructor and attendees run SAS codes that are provided in the workshop slides on two SAS practice datasets. Discuss the results as a group. 2) Students modify and run the SAS codes on their own data to examine the assumptions. Discuss their results with the instructor and other attendees.
diff --git a/lessons/assumptions/slides.md b/lessons/assumptions/slides.md
new file mode 100644
index 0000000..42c9624
--- /dev/null
+++ b/lessons/assumptions/slides.md
@@ -0,0 +1,219 @@
+---
+title: "Know your data and how to analyze it correctly: Statistical assumptions"
+date: 2015-02-13
+author: Daiva & Luke
+classoption: xcolor=dvipsnames
+output:
+ beamer_presentation:
+ slide_level: 1
+header-includes:
+ - \input{../slideOptions.tex}
+
+---
+
+# Welcome to our Statistical Assumptions workshop #
+
+## Purpose: ##
+
+To teach the statistical assumptions of linear regression and show how you test data to see if they satisfy the assumptions. Knowing how to check these assumptions is part of "best practices" in data analysis.
+
+
+## Significance: ##
+
+It is very important to check that your data satisfies linear regression assumptions. If your data does not meet these criteria, the use of linear regression is inappropriate. Other methods can be used, but...
+
+
+# Caveat (again): We aren't here to teach statistics #
+
+Need help with stats? Use these resources!
+
+* U of T Statistical Consulting Services ([click here](http://www.utstat.toronto.edu/wordpress/?page_id=25))
+
+*
+
+*
+
+* Helpful statistical tests flowchart (PDF on GitHub)
+
+* Very helpful webpage on regression diagnostics:
+
+
+# Notes and help during this workshop #
+
+* Go to this website:
+
+
+
+* Download our SAS code files from our GitHub page:
+
+([click here](https://github.com/codeasmanuscript/materials/assumptions))
+
+* Download the Statistical Tests Flowchart from our GitHub page:
+
+([click here](https://github.com/codeasmanuscript/materials/assumptions))
+
+# Linear Regression #
+
+* Used to test associations between independent and dependent variables
+
+* Based on a linear relationship: $y = X\beta + \varepsilon$
+ - y = dependent variable(s)
+ - $\beta$ = slope
+ - X = independent variable
+ - $\varepsilon$ = error, or residual, terms
+
+# Some Linear Regression Assumptions #
+
+* Model is good (i.e. linear relationship between predictors and outcome variable)
+
+* Residuals[^1] have a normal distribution
+
+* Residuals are homoscadastic (have equal/constant variance)
+
+[^1]: Residual (aka the error term) = observed - expected
+
+# Other Checks to Ensure Appropriate Model #
+
+* Check for collinearity (predictors that are highly linearly related -- may result in inaccurate estimates of regression coefficients)
+
+* Check for influence (i.e. outliers)
+
+# Brief aside: assumptions/diagnostics we are not covering in this workshop #
+
+* Independence (residuals of one observation are not associated with residuals of another)
+
+* Errors in variables (predictor variables are measured without error)
+
+* Very helpful webpage on regression diagnostics that covers these:
+
+# How to check assumptions #
+
+* Model fit: Make a scatterplot (check pattern)
+
+* Distribution of residuals: Q-Q Plot
+
+* Variance of residuals: Plot residuals vs. predicted fit (check spread of points)
+
+# Model fit #
+
+* Run a scatter plot:
+
+```
+
+ proc sgplot data=sashelp.class;
+ scatter x=height y=weight;
+ run;
+
+```
+
+# Model fit #
+
+
+
+# Residual distribution #
+
+ * Run a linear regression model and output the residual and predicted terms to a new dataset:
+
+```
+
+ proc reg data=sashelp.class;
+ model height=weight;
+ output out=resid residual=r predicted=fit;
+ run;
+ quit;
+
+```
+
+* Create a plot of the new output dataset:
+
+```
+ goptions reset=all;
+ proc univariate data=resid normal;
+ var r;
+ qqplot r / normal(mu=est sigma=est);
+ run;
+```
+
+# Residual distribution #
+
+
+
+# Residual variance #
+
+* Run a linear regression model and plot residuals against predicted values:
+
+```
+
+ proc reg data=sashelp.class;
+ model height=weight / spec;
+ plot r.*p.;
+ run;
+ quit;
+
+```
+
+# Residual variance #
+
+
+
+# What do you do if your data does not meet these assumptions? #
+
+> * Try transforming the data (log, square root)
+
+ data new;
+ set sashelp.fish;
+ logWt = log(Weight);
+ run;
+
+> * Use a non-parametric statistical test if can not obtain normal distribution of residuals after attempting a transformation
+
+
+# Collinearity #
+
+* What is it? Two or more predictors in a model that are moderately to highly correlated with one another (e.g. BMI and body weight)
+
+. . .
+
+* Check VIF (variance inflation factor)
+ - OR Check tol (tolerance = 1/vif)
+
+```
+
+ proc reg data=sashelp.class;
+ model height = weight age / vif tol;
+ run;
+ quit;
+
+```
+
+* VIF > 10 or tol < 0.1 suggest collinearity is present
+
+# Influence #
+
+> * Make a scatterplot of all observations
+```
+
+ proc sgplot data=sashelp.class;
+ scatter x=height y=weight;
+ run;
+```
+> Or another way to make a scatterplot:
+```
+
+ proc gplot data=sashelp.class;
+ plot height*weight=1 / vaxis=axis1;
+ run; quit;
+```
+> * Do a visual check for extreme observations
+
+# Influence cnt'd #
+
+> * Another method: proc univariate will output extreme observations
+
+> * Observation is "influential" if removing it substantially changes the estimate of coefficients (sometimes! exception: genetics--extreme observations may be hyper/hypo-responders)
+
+# Practice #
+
+1. Perform these checks on your own research data.
+2. Conclude if linear regression is appropriate and if collinearity or influence is present in your model.
+
diff --git a/lessons/git/assignment.md b/lessons/git/assignment.md
new file mode 100644
index 0000000..c0ccb2b
--- /dev/null
+++ b/lessons/git/assignment.md
@@ -0,0 +1,60 @@
+---
+title: "Assignment: Git"
+author:
+ - Luke W. Johnston
+date: 2015-10
+layout: page
+sidebar: false
+tag:
+ - Lessons
+ - Assignment
+ - Git
+categories:
+ - Lessons
+ - Git
+---
+
+## Challenges: Try these out yourself! ##
+
+In order to learn how to use Git, you really need to just start using it and get
+some practice! *Use only the commandline/terminal*. Don't forget to push them
+to your GitHub account and submitting a pull request at the end, so we can give
+feedback/make suggestions.
+
+Make sure to also have these folders in your own directory on the practice *Code
+As Manuscript* repo (called `practice-YYYY-MM` where `YYYY-MM` represents the
+year and month):
+
+ your-name/
+ - bio/
+ - git/
+ - intror/
+ - plotting/
+ - wrangling/
+ - rmarkdown/
+
+Now, make sure all of these challenges are placed in the proper folder (`git/`).
+
+1. Create a new file under the `your-name/git/` directory called `aboutme.txt`.
+ Write up a bit about yourself, some hobbies, what you are doing, etc. Now
+ get Git to track (add) that file and commit it to the history.
+2. Make a change to the `aboutme.txt` file by including something else about
+ yourself and making some edits to what you already wrote. Run `git diff` and
+ save the output of the `git diff` into a file called `diff.txt` (Hint: run
+ `git diff aboutme.txt > diff.txt`). **Don't commit the change yet**.
+3. Create a new file called `how-git-works.txt` and write up how you understand
+ how Git works, using your own words. After you have done that, do `git
+ status` and save the output into a file called `status.txt` (Hint: see the
+ hint in challenge 2 above). After you have created the `status.txt` file,
+ commit the files one by one, making sure to write up **descriptive** commit
+ messages!
+4. Add to the `how-git-works.txt` file and explain why you think Git and version
+ control are important. Then, add another file called `filesystem.txt` and
+ write up how *you* understand how filesystems work. Add and commit these
+ files to the git history, making sure to make *descriptive* commit messages.
+5. Lastly, push these files up to your GitHub account and submit a pull request
+ of your changes with the original practice Code As Manuscript
+ repository. Once pull requests have been completed, make sure to update your
+ local and your own GitHub repository with the original GitHub repository
+ (Hint: `git pull upstream master`).
+
diff --git a/lessons/git/cheatsheet.md b/lessons/git/cheatsheet.md
new file mode 100644
index 0000000..6f3ab78
--- /dev/null
+++ b/lessons/git/cheatsheet.md
@@ -0,0 +1,359 @@
+---
+title: "Cheatsheet: Git"
+author:
+ - Luke W. Johnston
+date: 2015-10
+layout: page
+sidebar: false
+tag:
+ - Lessons
+ - Cheatsheet
+ - Git
+categories:
+ - Lessons
+ - Git
+---
+
+Git is an amazing and very powerful tool that is useful for managing
+your projects and tools, letting you experiment and try out new things
+in your files without worrying about losing anything. However,
+because of its power and usefulness, it can be confusing for beginners
+as 1) Git requires the use of the command line (or terminal) to run
+Git commands, and 2) there are a large number of commands and options
+available. So, we put together the commands that are the most useful
+and most common --- and the only ones you may ever use! Also, please check out
+the [resources page](/lessons/resources) as it has links to very useful sites
+and tutorials on learning Git.
+
+As well, an added benefit of using Git is you can use the amazing
+[GitHub](https://github.com/), a git repository hosting service, to keep an
+online backup of your project and optionally share it with the world or with
+your team. There are increasing arguments and support for science to be more
+open and publicly accessible. GitHub can help with getting your code and/or
+data out into the public domain. Some of the below commands are used for
+communicating with GitHub, though they are not exclusive to GitHub.
+
+# Before using Git: Initial setup #
+
+This following commands should be run first before any work is done
+using Git.
+
+ git config --global user.name "Your Name"
+ git config --global user.email "you@some.domain"
+ git config --global color.ui "auto"
+ git config --global core.editor "your_editor"
+ git config --global push.default simple
+ git config --list
+
+These commands basically tell Git:
+
+* Who you are (`user.name`)
+* What your email address is (`user.email`; used when working on a
+ multiple person project)
+* What the colour output should be after running Git commands
+ (`color.ui`)
+* What your **text** editor is that you use (`core.editor`). See
+ [our instructions](/lessons/instructions/) for more
+ details about editors, but briefly:
+ - On Windows, you will likely use
+ [Notepad](http://en.wikipedia.org/wiki/Notepad_%28software%29)
+ (which comes pre-installed on Windows) or
+ [Notepad++](http://notepad-plus-plus.org/)
+ - On Mac, there is
+ [TextWrangler](http://www.macupdate.com/app/mac/11009/textwrangler)
+ (which I believe comes pre-installed) or
+ [TextMate](https://macromates.com/)
+ - On Linux, it depends on which distro you use, but in general
+ [gedit](https://help.ubuntu.com/community/gedit) or
+ [nano](https://wiki.archlinux.org/index.php/Nano) usually are
+ pre-installed
+* How you want the push and pull default behaviour (`push.default`;
+ more on this in the [GitHub lesson](/lessons/github/))
+
+# Terminal commands (good to know before knowing Git) #
+
+## Directory (terminology) ##
+
+> A directory is the same thing as a folder.
+
+## `cd path/to/directory` ##
+
+> Change the current directory/folder to another directory/folder. The little
+> tilde `~` means 'Home', so that would be `/home/username/` for Linux,
+> `/Users/username/` for Mac, and `C:\Users\username\` on Windows.
+
+> Example code:
+
+ cd ~/Desktop # Change to the desktop
+ cd /home/username/Documents/ # Change to documents.
+
+## `mkdir directory-name/` ##
+
+> Make (mk) a directory (dir) that has the name `directory-name`. This can be
+> any name you want it to be.
+
+> Example code:
+
+ mkdir ~/Desktop/testing # Create a testing folder on your desktop
+ mkdir playingAround/ # Make a folder in the current directory
+
+## `ls` ##
+
+> View the contents of the directory, showing files and sub-directories.
+
+> Example code:
+
+ ls ~/ # Show contents of home
+ ls -a ~/ # Show hidden and non-hidden files
+
+ # List files/directories by row, indicating which is a folder
+ # l = list format option
+ # h = human understandable bytes option
+ # F = indicate which are folders with a `/` at the end
+ ls -lhF ~/
+ ls -l -h -F ~/ # Same as above
+
+## `touch filename.txt` ##
+
+> Create an empty text file in the current directory called 'filename.txt'.
+
+> Example code:
+
+ # All these do the same thing.
+ touch filename.txt
+ touch File.txt
+ touch this-is-a-file.txt
+
+## `command > filename.txt` ##
+
+> Send the output into a file (in this case called 'filename.txt'.
+
+> Example code:
+
+ ls -lhF > filesInDirectory.txt # Send ls output to file
+ echo 'This is a test' > testing.txt # Send to the file
+
+
+# Useful (and common) Git commands #
+
+## Repository (terminology) ##
+
+> Not a command, but is the term used to describe all the saved history of a
+> directory and files for a project that are tracked by Git. Is essentially the
+> hidden `.git/` directory.
+
+## `git init` ##
+
+> Tell Git to start tracking a folder by creating a git history
+> repository. This essentially tells Git to start watching your
+> folder and all the files and folders within it.
+
+> Example code:
+
+ ## Comment: cd = change directory (aka folder)
+ cd ~/Documents/yourprojectname/
+ git init
+
+## `git status` ##
+
+> Tell Git to list out all the activity within a folder under watch by
+> Git (after using `git init`). The status command will list all
+> files or folders that have been added or changed inside the
+> repository.
+
+> Example code:
+
+ cd ~/Documents/yourprojectname/
+ git status
+
+## `git add ` ##
+
+> Add the files you want Git to watch inside the history repository as
+> created with `git init`. You can specify as many or as little files
+> or folders as you want. *Note*: the add command does **not** save
+> the files to the git history. All the `git add` command does is
+> tell Git to start watching the files and put the files into the
+> "staging area" where they will next be saved to the history (see the
+> below `git commit` command).
+
+> Example code:
+
+ ## Comment: cd to where files have been changed
+ cd ~/Documents/yourprojectname/
+ ## Comment: Pretending we want to add three files
+ git add foldername/newfilename1 newfilename2 foldername2/changedfile
+
+## `git commit` ##
+
+> Tells Git to save your `git add`'ed files to its history. This is
+> the main purpose and use for Git. After you commit a file(s) and
+> typed out a commit message (*be detailed about what you did*!!), Git
+> will save (or take a "snap shot") of your files and put it into its
+> history. Once committed, it is saved into the history, allowing you
+> to go back to that commit/save point at any time!
+
+> Example code:
+
+ cd ~/Documents/yourprojectname/
+ ## Comment: You have two options...
+ ## Commit and let a text editor pop up so you can
+ ## write your commit message
+ git commit
+ ## Comment: ... Or you can use the -m option
+ git commit -m "Type out your commit message here"
+
+## `git log` ##
+
+> Displays the history of your repository and the messages you added
+> to each commit, as well as the date of the change, and who made the
+> change. This is analogous to a logbook for those in the basic
+> science. This is a really useful feature if you are working on a
+> multi-person project or if you are coming back to a project after
+> several months of not touching it and completely forgetting what you
+> were doing last. `git log` has a *large* number of options
+> available that customize the appearance and the information provided
+> by the log command.
+
+> Example code:
+
+ cd ~/Documents/yourprojectname/
+ git log
+
+## `git checkout` ##
+
+> Allows you to go back (or forward) throughout your Git history as
+> well as to change branches (see the `git branch` command below). If
+> you wanted to go back a few commits, you would run `git checkout
+> commitnumber`. You get the commit number by running git log and
+> using the first few letters and numbers of the commit. We will
+> likely show an example in the workshop.
+
+> Example code:
+
+ ## Comment: Go back to a previous commit (use the commit hash)
+ git checkout d45gfd3 ## Example commit hash, found using git log
+ ## Comment: Go to another branch (ie: "testing")
+ git checkout testing
+ ## Comment: Go back to your main branch ("master")
+ git checkout master
+
+## `git branch ` ##
+
+> This is another very useful command from Git, and (I feel) one of
+> its biggest strengths! This command basically makes a branch like
+> on a tree, letting you experiment with your files and statistical
+> analyses without having to worry about messing around with your main
+> files. If you want to eventually bring the experimental branch into
+> the "master" branch, you can use
+> [`git merge`](http://git-scm.com/book/en/v2/Git-Branching-Basic-Branching-and-Merging)
+> (which we will not be covering as it is a slightly more advanced
+> command).
+
+> Example code:
+
+ ## Create a branch named "experiment"
+ git branch experiment
+ ## Move to the experiment branch
+ git checkout experiment
+ ## edit your files and save, do whatever...
+ git add changedfiles
+ git commit -m "Added a statistical test with different variables"
+ ## Move back to the master branch
+ git checkout master
+ ## All files should be in their original state
+
+## `git diff ` ##
+
+> This command tells Git to compare the contents of, at the basic
+> level, two files. These two files are by default a file you most
+> recently changed (but not committed) with the same file's content in
+> the history. However, you can also compare a file across different
+> commits in the history. `git diff` shows the differences between
+> the files by highlighting in red deletions and in green additions to
+> the file. This is useful if you forgot what you changed in the file
+> and you are going to commit the file into the history, using it to
+> help you write a better commit messages.
+
+> Example code:
+
+ ## Compare a recently changed file with its recent
+ ## commit history
+ git diff filename
+ ## Compare a file across two commits (using commit hashes)
+ ## Usage: git diff hash1..hash2 file (hashes are *very* unique)
+ git diff 54gfd..75g84 filename
+
+# Git commands for dealing with GitHub/other hosting services #
+
+## `git clone ` ##
+
+> Cloning is basically downloading a new git repository that is
+> online/in GitHub. You take an existing git repository and
+> duplicate/copy/clone it onto your own computer. For example, if you
+> wanted the files for this workshop series, you would first fork our
+> [GitHub Code As Manuscript workshop repo](https://github.com/codeasmanuscript/workshops)
+> so you have your own copy of it on your account and then you clone
+> it into your computer.
+
+> Example code:
+
+ ## First fork our Code As Manuscript and then:
+ cd /path/to/where/you/want/the/repo
+ git clone https://github.com/your-name/workshops.git
+ ## You now have the workshop files on your computer.
+
+## `git remote add ` ##
+
+> A remote, in git terminology, is a server or online location.
+> Remote is the opposite of local. A remote repository is a
+> repository that is *not* on your computer, while a local repository
+> is. Think of the remote as an external hard drive. When you add a
+> remote, you use a name (by convention the name is usually "origin",
+> which I *strongly* encourage you to use as well) that will tell git
+> that that is the remote name. The server url can generally be found
+> on the GitHub or BitBucket page, usually in the top or bottom right
+> corner. For instance, the server url for my own 'test' project
+> would be `https://github.com/lwjohnst86/test.git`. to describe your
+> Git project that you want to store on the server such as Github.
+
+> Example code:
+
+ cd /path/to/your/git/repo
+ ## The actual URL can be found on GitHub, usually in the corner.
+ git remote add origin https://github.com/yourname/yourproject.git
+ git push
+
+## `git push` ##
+
+> Push is essentially the same as uploading your local git repository
+> to the remote (GitHub) repository. Pushing is more powerful than a
+> simple upload, as git checks the remote repository, compares it to
+> the local repository, making sure that the integrity of the files is
+> preserved and that nothing is lost or overwritten (unlike Dropbox
+> for example). This is especially important for your research files!
+
+> Example code:
+
+ cd /path/to/your/git/repo
+ ## ... edit a file ...
+ git commit -am "Edited a file"
+ git push
+
+## `git pull` ##
+
+> This is essentially a command to download the remote (GitHub)
+> repository contents and merge it into your own local git repository.
+> This is only ever used if you a) work on a project that is only more
+> than one computer and you use git and GitHub to sync the files
+> across computers, or b) if you work on a team and one or more other
+> person(s) are making changes to the remote (GitHub) repository.
+> Pulling then syncs the updated remote content with your local
+> content. This is where git really shines when you collaborate with
+> others on a project (ie: this workshop series)!
+
+> Example code:
+
+ cd /path/to/your/git/repo
+ ## Someone has added stuff to the remote repo
+ git pull
+
diff --git a/lessons/git/intro.md b/lessons/git/intro.md
new file mode 100644
index 0000000..9872765
--- /dev/null
+++ b/lessons/git/intro.md
@@ -0,0 +1,237 @@
+---
+title: "Introduction: Version control using Git and GitHub"
+author:
+ - Luke W. Johnston
+ - Daiva Nielsen
+date: 2015-10
+layout: page
+sidebar: false
+tag:
+ - Lessons
+ - Introduction
+ - Git
+categories:
+ - Lessons
+ - Git
+permalink: lessons/git/
+---
+
+Version control is a system that manages changes to a file or files.
+These changes are kept as logs in a history, with detailed information
+on what file(s) was changed, what was changed within the file, who
+changed it, and a message on why the change was made. This is
+extremely useful, especially when working in teams or for yourself 6
+months in the future (because you *will* forget things)!
+
+To understand how incredibly powerful version control is, think about
+these questions: How many files of different versions of a manuscript
+or thesis do you have laying around after getting feedback from your
+supervisor or co-authors? Have you ever wanted to experiment with your
+code or your manuscript and need to make a new file so that the
+original is not touched? Have you ever deleted something and wish you
+hadn't? Have you ever forgotten what you were doing on a project?
+
+Version control fixes *all* of these problems! And our aim for this
+workshop is to get you familiar with Git and (at least partly)
+comfortable with the concept.
+
+## Learning expectations ##
+
+After this workshop, our expectation is that you will:
+
+- Know how to use the basic tools of version control in your work
+- Know how to collaborate with others on a project
+- Have a basic understanding of how Git and GitHub works
+- *Know where to go for help*
+
+# Materials for this lesson: #
+
+* [Slides](slides/)
+* [Cheatsheet](cheatsheet/)
+* [Assignment](assignment/)
+
+Other resources can be found [here](/lessons/resources/)
+
+# Brief aside: Filenaming #
+
+Because Git keeps a record of your files and changes to them, you
+don't need to have different files names for different versions
+(eg. file-V01.doc, file-V02.doc, etc). Plus, because part of the
+benefit of Git is eventually being able to collaborate, it's
+*extremely* useful to follow some rules on filenaming. We've put
+together some below:
+
+1. Keep the names short, but meaningful. Remove unnecessary words such
+ as "the", "and", "a" etc.
+
+2. Don't include spaces and avoid underscores (debatable and/or
+ situational). For a string of words, capitalize the first letter
+ of each word, except for the first word
+ (e.g. `fileNameDescription`)
+
+3. Use hyphens to separate important parts of the name or when there
+ is an abbreviation followed by another word. For example,
+ `diabetesRisk-AnalysisOutput` which separates the two concepts, the
+ project descriptor `diabetesRisk` and the contents of the file
+ `AnalysisOutput`. Another example:
+ `report-AnalysisFoodIntake-2014.pdf` and **not**
+ `reportAnalysisFoodIntake2014.pdf`.
+
+4. Avoid redundancy in file names and file paths (folder names). For
+ example, don't use `folderName/fileName-folderName.txt` and instead
+ use `folderName/fileName.txt`. Another example: **don't** use
+ `diabetesFats/analysis-DiabetesFats.sas` and **instead** use
+ `diabetesFats/analysis.sas`.
+
+5. If a number is included in the filename, such as for the version
+ number, use two digits not one (e.g. `V01`, not `V1`).
+
+6. When including a date, include it at the very end of the filename
+ and in the international standard format `YYYY-MM-DD` (in all numbers).
+
+# Brief aside 2: Tips on using the terminal #
+
+- Make use of TAB-completion in the terminal!
+- Up arrow on the terminal goes to the previous command you entered.
+
+# Putting your files under version control using Git #
+
+## Setting up your user information and GitHub account ##
+
+For configuring your git, follow the
+["Initial setup" in the cheatsheet page](http://codeasmanuscript.org/lessons/git/cheatsheet/)
+I've put together.
+
+Then, set up a [GitHub](https://github.com/) account if you haven't already. If
+you are concerned about having your research or data be public you can either a)
+[request](https://education.github.com/) 5 free private repos from GitHub or b)
+use [BitBucket](https://bitbucket.org/), which uses private repos by
+default. But at least for this workshop, set up a GitHub account.
+
+## Basics of using Git for a fresh repository ##
+
+Create a folder and create a git repository (which is the stored
+history) in that folder. (Note: `##` is a comment, not a command).
+
+ cd ~/Desktop ## Move to your desktop
+ mkdir playing ## Create a folder (aka directory)
+ cd playing
+ git init ## Create the repository (init = initialize)
+
+Now, create a file, get git to track it, and save it to the history.
+
+ touch bio.txt ## Command to create a file called bio.txt
+ ls ## Check that you created the file, ls = list files
+ git add bio.txt ## Track the file
+ ## Save the file to the history with a message (-m)
+ git commit -m "Initial commit"
+
+Now, open the `bio.txt` file and add:
+
+* Your name
+* Your program and year
+* Your progamming language/statistical language of choice
+
+Then:
+
+ git status ## Check the activity
+ git diff bio.txt ## Compare to the one in the history
+ git add bio.txt ## This sends it to the staging area
+ git commit -m "Added my bio" ## This sends it to the history
+
+For a description on what the different stages are (working directory,
+staging area, and committed history) see the below links or check out the
+[resources page](/lessons/resources/):
+
+* Description: https://git-scm.com/book/en/v2/Getting-Started-Git-Basics
+* Image: https://git-scm.com/book/en/v2/book/01-introduction/images/areas.png
+
+Then, to see what has happened in your history:
+
+ git log ## View the log of your history
+ git log --graph --oneline ## A condensed view of the log
+
+## Including GitHub in your workflow ##
+
+One of the major strengths of Git are it's features that allow easy
+collaboration on complex (or simple) projects. GitHub, which is a hosting
+service (not a program and not related to Git), uses Git to save and share Git
+repositories online. Most, if not all of the open source community has their
+projects on GitHub. You could set up your own GitHub repository, but for now, we
+will ['fork'](https://help.github.com/articles/fork-a-repo/) (which means copy
+someone else's GitHub repository in GitHub language) the *Code as Manuscript*
+practice repository (in the form `practice-YYYY-MM`). There is a button in the
+top right corner called 'Fork' that allows you to do that. Click it. The
+`practice-YYYY-MM` repository is now on your GitHub account! Now we can 'clone'
+(meaning download the git repository onto your computer) your newly created
+'forked' repository!
+
+ mkdir ~/Desktop/workshops/
+ cd ~/Desktop/workshops/
+ git clone https://github.com/yourusername/practice-YYYY-MM.git
+ cd practice-YYYY-MM/
+
+Now it should have downloaded the repository onto your computer. Because you
+cloned forked repository, it's good practice to also include a link to the
+original (upstream) repository:
+
+
+ git remote add upstream https://github.com/codeasmanuscript/practice-YYYY-MM.git
+
+You can view the different links (called remotes) you have to your GitHub
+accounts:
+
+ git remote -v # v = verbose, or rather more detailed
+
+You can see you should have two remotes, `origin` (*your* forked repo) and
+`upstream` (codeasmanuscript repo). Now, create a new directory that you will
+use for the rest of the workshops, as well as all the directories you will use
+for the other workshop topics:
+
+ mkdir your-name/
+ cd your-name/
+ # Create multiple directories by typing in more names
+ mkdir git intror plotting wrangling rmarkdown
+ cd git/ # Move into the git one
+
+Now, create a `bio.txt` file, write up your name and program. Add and commit
+that file. Now we want to upload (`push`) your new file onto your GitHub repo
+and submit a
+['pull request'](https://help.github.com/articles/using-pull-requests/):
+
+ git push origin master # Origin is your account
+
+Go to GitHub and click the 'Pull Requests' button on the side and on the next
+page click the 'New pull request' button. Keep clicking until you have finished
+submitting your pull request. After we (the *Code As Manuscript* instructors)
+accepted the pull requests, you need to update your local git repo and your
+forked GitHub repo by downloading (`pull`) from the original `upstream` GitHub
+repo.
+
+ git pull upstream master
+ git push origin master
+
+That covers the basics of using Git and incorporating GitHub into the workflow!
+If you ever need help with any commands, check out
+[StackOverflow](http://stackoverflow.com/) for answers on tens of thousands of
+questions with tens of thousands of answers!
+
+# Brief glossary of terms: #
+
+* `cd` - change directory
+* directory - the same thing as a folder
+* `ls` - list the files and folders in a folder
+* `touch` - create an empty file
+* repository - the saved history of a folder and files, used by git and located
+ in the `.git/` folder.
+* `init` - start or initialize a git repository
+* `add` - put a file into the staging area, so that git starts
+ tracking it
+* staging/index area - where files are stored before going into the
+ history
+* `commit` - send files in the staging/index area into the history
+ (the git repository)
+* `status` - check the status of the folder and the git repository
+* `diff` - compare a file to the a file in the history
+* `log` - view the commit history in the git repository
+
diff --git a/lessons/git/slides.md b/lessons/git/slides.md
new file mode 100644
index 0000000..d138f6e
--- /dev/null
+++ b/lessons/git/slides.md
@@ -0,0 +1,72 @@
+---
+title: "Let's 'Git' started: Using version control"
+author: Luke W. Johnston
+date: 2015-10
+layout: page
+sidebar: false
+tag:
+ - Lessons
+ - Slides
+ - Git
+categories:
+ - Lessons
+ - Git
+output: slidy_presentation
+---
+
+##
+
+
+
+## Learning expectations ##
+
+- How to use version control for your work
+- How to collaborate with others on a project
+- Have a basic understanding of how Git and GitHub works
+- *Know where to go for help*
+
+## 5 main concepts ##
+
+- **Start repository**: git init, git clone (GitHub)
+- **Check activity**: git status, git log, git diff
+- **Save to history**: git add, git commit
+- **Move through the history**: git checkout
+- **Synchronizing with GitHub**: git push, git pull
+
+## Visualization of how Git saves images ##
+
+](/lessons/images/vcs-Snapshot.png)
+
+## Actual visualization of Git history ##
+
+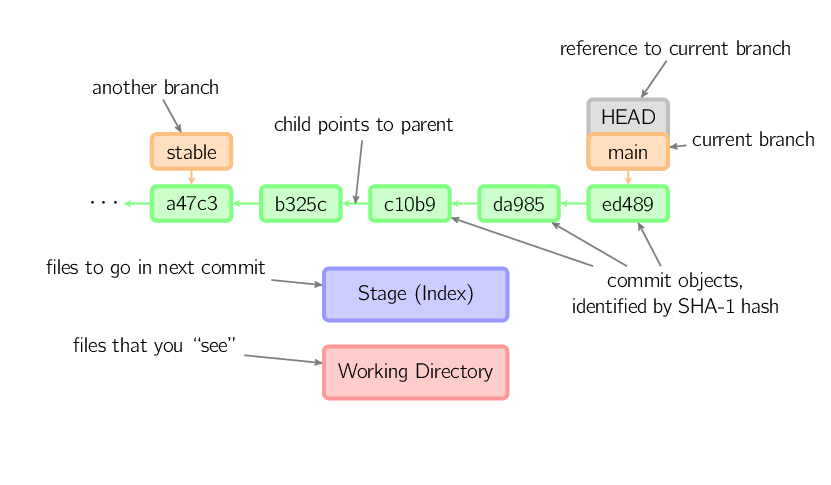
+
+## Different areas Git uses ##
+
+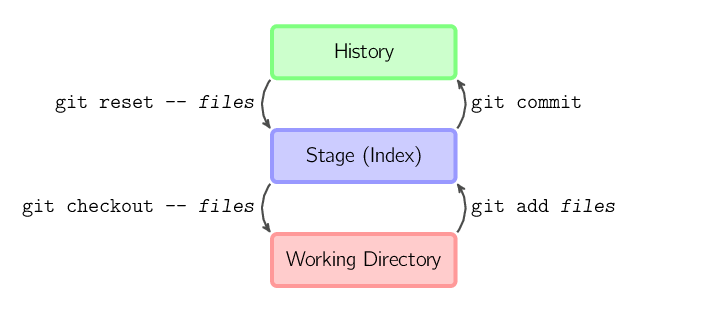
+
+## Configuring Git ##
+
+```
+git config --global user.name "Your Name"
+git config --global user.email "you@some.domain"
+git config --global color.ui "auto"
+git config --global core.editor "your_editor"
+git config --global push.default current
+git config --list
+```
+
+## How GitHub works ##
+
+
+
+## How GitHub Forking (collaboration) works ##
+
+
+
+## Generate these slides using (using R): ##
+
+
+{% highlight r %}
+rmarkdown::render('slides.Rmd')
+{% endhighlight %}
diff --git a/lessons/images/GitHubFlow.png b/lessons/images/GitHubFlow.png
new file mode 100644
index 0000000..d6254ac
Binary files /dev/null and b/lessons/images/GitHubFlow.png differ
diff --git a/lessons/images/GitHubForkFlow.png b/lessons/images/GitHubForkFlow.png
new file mode 100644
index 0000000..f66f051
Binary files /dev/null and b/lessons/images/GitHubForkFlow.png differ
diff --git a/lessons/images/PLOS.png b/lessons/images/PLOS.png
new file mode 100644
index 0000000..4783b23
Binary files /dev/null and b/lessons/images/PLOS.png differ
diff --git a/lessons/images/PLOSdate.PNG b/lessons/images/PLOSdate.PNG
new file mode 100644
index 0000000..8615aab
Binary files /dev/null and b/lessons/images/PLOSdate.PNG differ
diff --git a/lessons/images/fig-1.png b/lessons/images/fig-1.png
new file mode 100644
index 0000000..9324a2c
Binary files /dev/null and b/lessons/images/fig-1.png differ
diff --git a/lessons/images/filenamingComic.gif b/lessons/images/filenamingComic.gif
new file mode 100644
index 0000000..721323e
Binary files /dev/null and b/lessons/images/filenamingComic.gif differ
diff --git a/lessons/images/gitBranches.png b/lessons/images/gitBranches.png
new file mode 100644
index 0000000..3762e6c
Binary files /dev/null and b/lessons/images/gitBranches.png differ
diff --git a/lessons/images/modelFit.jpg b/lessons/images/modelFit.jpg
new file mode 100644
index 0000000..1361643
Binary files /dev/null and b/lessons/images/modelFit.jpg differ
diff --git a/lessons/images/plotCap-1.png b/lessons/images/plotCap-1.png
new file mode 100644
index 0000000..5acb6c0
Binary files /dev/null and b/lessons/images/plotCap-1.png differ
diff --git a/lessons/images/raw/GitHubFlow.dia b/lessons/images/raw/GitHubFlow.dia
new file mode 100644
index 0000000..fedf7f2
Binary files /dev/null and b/lessons/images/raw/GitHubFlow.dia differ
diff --git a/lessons/images/raw/GitHubForkFlow.dia b/lessons/images/raw/GitHubForkFlow.dia
new file mode 100644
index 0000000..a46af4f
Binary files /dev/null and b/lessons/images/raw/GitHubForkFlow.dia differ
diff --git a/lessons/images/reproducibleCrisis.png b/lessons/images/reproducibleCrisis.png
new file mode 100644
index 0000000..fb0862f
Binary files /dev/null and b/lessons/images/reproducibleCrisis.png differ
diff --git a/lessons/images/residNorm.jpg b/lessons/images/residNorm.jpg
new file mode 100644
index 0000000..fb06d96
Binary files /dev/null and b/lessons/images/residNorm.jpg differ
diff --git a/lessons/images/residVar.jpg b/lessons/images/residVar.jpg
new file mode 100644
index 0000000..35ad371
Binary files /dev/null and b/lessons/images/residVar.jpg differ
diff --git a/lessons/images/vcs-Snapshot.png b/lessons/images/vcs-Snapshot.png
new file mode 100644
index 0000000..f6d9162
Binary files /dev/null and b/lessons/images/vcs-Snapshot.png differ
diff --git a/lessons/instructions.md b/lessons/instructions.md
new file mode 100644
index 0000000..a1eef8a
--- /dev/null
+++ b/lessons/instructions.md
@@ -0,0 +1,71 @@
+---
+layout: page
+sidebar: false
+title: Pre-workshop instructions
+permalink: lessons/instructions/
+---
+
+Before coming to any of our workshops, please make sure to do the
+following:
+
+* Register (either from our email or from our through GPS)
+* Install the required software (Git, SAS, or R) on your laptop
+* Make sure to fully charge your laptop
+
+# Software installation #
+
+Please install these programs before coming to the workshops. If you
+have issues with installing the programs, please come 15 minutes early
+to the workshop and we'll try to help you out.
+
+## Git ##
+
+Installation instructions for Git can be found the
+[Git download site](http://git-scm.com/book/en/Getting-Started-Installing-Git)
+
+* On Windows: Download the `.exe` file from this
+ [Github link](http://msysgit.github.io) and run it.
+
+* On Mac: To use the graphical Git installer, download from this
+ [SourceForge link](http://sourceforge.net/projects/git-osx-installer/)
+
+* On Linux: Use your respective package manager (for example, if you
+ use Ubuntu or Debian, run this code: `sudo apt-get install git`)
+
+## Text editor ##
+
+Because we will be writing plain text (for
+[Markdown](https://guides.github.com/features/mastering-markdown/) and
+Git), you'll need a text editor. Most operating systems (Microsoft,
+Mac, Linux) generally come pre-installed with a text editor.
+
+* Windows: Notepad is installed by default, however
+ [Notepad++](http://notepad-plus-plus.org/download/v6.6.9.html) is
+ really good. Another option is
+ [MarkdownPad](http://markdownpad.com/).
+
+* Mac: [TextMate](http://macromates.com/download) is good, check the
+ website to install it.
+ [Nano](http://en.wikipedia.org/wiki/GNU_nano) is also (usually)
+ pre-installed.
+
+* Linux: Most distributions have pretty good pre-installed plain text
+ editors. For instance, Ubuntu and Debian have gedit. Nano is also
+ generally pre-installed.
+
+## SAS (for our SAS workshops) ##
+
+The installation instructions for SAS tend to be not the most
+intuitive (especially for Linux users). Mac users can not use SAS
+natively and must dual-boot onto Windows. SAS has their own
+installation guide that comes with the CDs.
+
+## R (for our R workshops) ##
+
+To install R, go to the [R Project website](http://cran.rstudio.com/)
+and choose your operating system. Unlike SAS, R does not come with an
+editor, so you will need to install an editor. One very useful and
+beginner-friendly editor is [RStudio](http://www.rstudio.com). For
+more advanced users who are eager for a (substantial) challenge, you
+can try [Emacs and ESS](http://ess.r-project.org/).
+
diff --git a/lessons/lessons.md b/lessons/lessons.md
new file mode 100644
index 0000000..3aa5164
--- /dev/null
+++ b/lessons/lessons.md
@@ -0,0 +1,49 @@
+---
+layout: page
+sidebar: true
+title: Lessons
+permalink: lessons/
+---
+
+> Hello and welcome to our Code As Manuscript site for data analysis
+> and coding-related workshops! This page contains links to
+> pre-workshop instructions, lesson material, and our teaching goals.
+
+# Instructions #
+
+Please check our [instructions](instructions/) before coming
+to the workshops. There will be instructions on what to install and
+how to install it. We **strongly** encourage you to do this *before*
+coming to the workshops as doing this during the workshops will delay
+the planned workshop material and slow everyone else down. Given
+that, there are times when you may run into difficults while
+installing something. In those cases, we will be at the workshop
+20-30 minutes beforehand and if you need any assistance, *please*
+come then and we will help out as much as we can!
+
+# Lessons #
+
+Our four primary workshops include learning about Git, GitHub, SAS
+macros, and SAS ODS. We hope to eventually develop a series that
+focuses on R, however, given that a large majority of (at least
+biomedical) researchers use SAS, we've developed the SAS workshops
+first.
+
+Our lesson material:
+
+## Our lesson material: ##
+
+{% for cat in site.lesson-list %}
+
+### {{ cat }}: ###
+
+
+ {% for page in site.pages %}
+ {% for pc in page.categories %}
+ {% if pc == cat %}
+ - {{ page.title }}
+ {% endif %}
+ {% endfor %}
+ {% endfor %}
+
+{% endfor %}
diff --git a/lessons/macros/assignment.md b/lessons/macros/assignment.md
new file mode 100644
index 0000000..67a2889
--- /dev/null
+++ b/lessons/macros/assignment.md
@@ -0,0 +1,39 @@
+---
+title: "Assignment: SAS Macros"
+author:
+ - Luke
+ - Daiva
+date: 2015-03-20
+fontsize: 12pt
+geometry: margin=1in
+papersize: letterpaper
+layout: page
+sidebar: false
+tag:
+ - Lessons
+ - Assignment
+ - Macros
+categories:
+ - Lessons
+ - Macros
+---
+
+Depending on your circumstances, there are two possibilities for practice:
+
+## Working solo ##
+
+* Make a repetitive code without using a macro. You can work with one of the SAS practice datasets if you don't have your own data. Track this file using Git.
+* Improve this code by making a macro. You can include both codes in the same .sas file. If you want you can even delete the long-style of code and do a "git diff" to see the changes to the file.
+* Give us some "Thoughts" about this workshop as a separate text file. Feel free to include points on how you think you can apply macros in your research.
+* Push both your SAS file and Thoughts file to the appropriate location in the Sandbox repo on GitHub.
+
+
+## Working in pairs ##
+
+Pair up with a peer and:
+
+* Each of you create your own file of a repetitive statistical code without using macros. Use the SAS practice datasets if you need to and track these files using Git.
+* Push both of your files to your personal Github repo.
+* Pull your partner's code file.
+* Apply macros to your partner's code to improve it.
+* Push to your personal Github repo and create a PR to merge to your partner's repo.
diff --git a/lessons/macros/cheatsheet.md b/lessons/macros/cheatsheet.md
new file mode 100644
index 0000000..c397d29
--- /dev/null
+++ b/lessons/macros/cheatsheet.md
@@ -0,0 +1,167 @@
+---
+title: "Cheatsheet: SAS Macros"
+author:
+ - Luke Johnston
+date: 2015-04-01
+fontsize: 12pt
+geometry: margin=1in
+papersize: letterpaper
+layout: page
+sidebar: false
+tag:
+ - Lessons
+ - Cheatsheet
+ - Macros
+categories:
+ - Lessons
+ - Macros
+---
+
+SAS has a powerful feature know as the macro language. If you have
+repetitive code, or a particular analysis that is fairly complex,
+macros are there to make your life easier! Below are some basic
+things to remember and to know for using macros. Luke also has a
+brief intro/tutorial on writing your own macros at
+[his blog](http://lwjohnst86.github.io/Introduction-Creating-Macro-SAS/).
+Luke also has developed a
+[personal macro library on GitHub](https://github.com/lwjohnst86/sasToolkit/src)
+that is fairly well documented, so you can look them over if you want.
+Maybe you will find something that suits your own analysis!
+
+# SAS macro commands: #
+
+## `%macro name (arg1, arg2=);` ##
+
+> This is the command format you would use to start a macro. An
+> example is shown in the "Example" section at the bottom.
+
+* `%macro` part tells SAS that the upcoming code is a macro.
+* `name` is the name you would give your macro, for example `means` or
+ `corr` or `regression` and so on.
+* `arg1` and `arg2` are known as arguments. They are used to include
+ other variables within the macro. A better explanation is below in
+ the example section.
+* `arg1` is known as a positional argument (or parameter) because it
+ has no `=` sign after it. A positional argument means that what
+ ever variable is first supplied to the macro takes on the value of
+ `arg1`. For example, for a macro such as `%macro means(vars,
+ where=);`, when you call the macro `%means(Height, where = Wgt <
+ 100);` the 'Height' variable takes on the value of `arg1` because it
+ is first.
+* `arg2=` is known as a keyword argument (or parameter) because of the
+ `=` sign. Thus, in order to use this argument, you need to
+ specifically call it. For example, `%means(Height, where = Wgt <
+ 100);` the `where` argument needs to be called directly, while
+ the `vars` argument is replaced by 'Height' because it is a
+ positional argument.
+
+## `%mend name;` ##
+
+> This ends the macro definition (`mend` = macro end). So to end the
+> `%macro means();` example, you use `%mend means;`. See the example
+> below.
+
+## `%let variable = something;` ##
+
+> This is known as a macro variable. The `%let` statement is kind of
+> like telling SAS to create a jar. You name this jar as 'variable'
+> and inside the jar you place 'something'. This can be very useful
+> when you have a long list of variables that you repeated use.
+
+> Example: `%let jar = BMI Wgt Hgt Age;`, 'jar' now contains
+> these 4 variables, which can be called using `&jar` (see below).
+
+## `&variable` ##
+
+> This is also known as a macro variable. However, unlike the `%let`
+> command above, here you are not creating a macro variable, but
+> rather telling SAS to use the contents of the macro variable from
+> the `%let` command (when you created the 'jar'). Continuing with
+> from the example directly above, `&jar` is replaced with 'BMI Wgt
+> Hgt Age' before SAS processes the `proc` or `data` command. Again,
+> see the example at the bottom.
+
+## `%if ... %then ...;` ##
+
+> This is known as a conditional. This is a fairly advanced component
+> of macros, but is really where using macros really starts to shine.
+> They let you expand your macro to include other components of code
+> without creating a whole new macro. Depending on time, we may or
+> may not cover this.
+
+## `%do i = 1 %to num;` ##
+
+> This is known as a 'do loop'. Like the `%if ... %then ...;` above,
+> this is an advanced but *extremely* powerful feature of macros that
+> lets you do some very impressive things! Given the advanced nature
+> of this command, we won't likely be going over this, but it's good
+> to know other features to use in macros.
+
+# Example macro: #
+
+We want to create a macro for calculating means, than running it on
+some some data. This is real code that can be run, so try it out on
+your own!
+
+ %macro means(vars, where=, class=, data=);
+ proc means data=&data;
+ var &vars;
+ where &where;
+ class &class;
+ run;
+ %mend means;
+
+ %let length = Length1 Length2 Length3;
+ %let others = Weight Height Width;
+
+ %means(&length, where = Weight < 200,
+ class = Species, data = sashelp.fish);
+
+ %means(&others, class = Species,
+ data = sashelp.fish);
+
+ %means(&length, data = sashelp.fish);
+
+## Lets break this macro down: ##
+
+ %macro means(vars, where=, class=, data=);
+
+This creates a macro called `means` that has 4 arguments, 1 of which
+is positional (`vars`).
+
+ proc means data=&data;
+ var &vars;
+ where &where;
+ class &class;
+ run;
+
+This is the meat of the macro. Using the ampersand `&`, we can place
+the arguments at various places throughout the macro. When SAS runs
+this code, `&data` will be replaced by what ever you put into it, and
+so on.
+
+ %mend means;
+
+This tells SAS that your own custom macro is finished.
+
+ %let length = Length1 Length2 Length3;
+ %let others = Weight Height Width;
+
+These two commands are macro variables. Basically, we are creating
+two 'jars' here, named 'length' and 'others'. Each 'jar' contains 3
+variables each.
+
+ %means(&length, where = Weight < 200,
+ class = Species, data = sashelp.fish);
+
+This is where we actually invoke the macro `means` that we created.
+Because `vars` was a positional argument, we don't have to call it
+directly (ie: `vars = &length`). Just putting `&length` in the first
+position tells SAS what the variable is. Because the other 3
+arguments were keyword arguments, they have to be explicitly called
+(eg: `where =`).
+
+This is a *very* basic example. They can get fairly complex, but
+*very* powerful as you add more components to the macro. Anytime you
+have repetitive or complex code, create a macro and recycle your
+code. This saves an incredible amount of time and headache!
diff --git a/lessons/macros/intro.md b/lessons/macros/intro.md
new file mode 100644
index 0000000..f9d2efb
--- /dev/null
+++ b/lessons/macros/intro.md
@@ -0,0 +1,69 @@
+---
+title: "Introduction: SAS Macros"
+author:
+ - Luke Johnston
+date: 2015-04-01
+fontsize: 12pt
+geometry: margin=1in
+papersize: letterpaper
+layout: page
+sidebar: false
+tag:
+ - Lessons
+ - Introduction
+ - Macros
+categories:
+ - Lessons
+ - Macros
+permalink: lessons/macros/
+---
+
+**In development**
+
+* Introductions
+* State goal for workshop
+* Item 2
+
+* Introduction
+ * Make sure they have git and github
+ * Clean code, code as manuscript, sanity
+ * What are macros
+ * How they are useful
+ * Steps-by-step to creating them
+ * General rules of thumb for making them
+ * Loops
+ * Best practices
+
+* Examples:
+ * Bad code, good code
+ * Daiva make bad code for means
+ * Luke make bad code for glm (anova)
+ * Both fix it (pre-WS)
+ * Example of my macro library
+ * Work through proc corr, proc glm, proc means
+ * Create their own of a test they use often
+ * Use their own data/analysis
+ * Or just make up their own
+
+* For future: mention power of combining with ODS
+
+## Learning objectives ##
+
+Our expectations are that after this workshop you
+
+1. Can explain fairly well what macros and macro variables are (how
+ they work, what the basic structure is, and how they are used).
+
+
+
+# Materials for this lesson: #
+
+* [Slides](slides/index.html)
+* [Cheatsheet](cheatsheet/index.html)
+* [Assignment](assignment/index.html)
+
+# Other resources: #
+
+* [link](http://)
+* [Luke's intro blog to SAS macros](http://lwjohnst86.github.io/Introduction-Creating-Macro-SAS/).
+* [Luke's personal macro library on GitHub](https://github.com/lwjohnst86/sasToolkit/src)
diff --git a/lessons/macros/slides.md b/lessons/macros/slides.md
new file mode 100644
index 0000000..25a8298
--- /dev/null
+++ b/lessons/macros/slides.md
@@ -0,0 +1,162 @@
+---
+title: "Fighting chaos: Tricks to re-use code and become more
+ productive"
+author: Daiva & Luke
+date: 2015-06-29
+layout: page
+sidebar: false
+classoption: xcolor=dvipsnames
+highlight-style: kate
+tag:
+ - Lessons
+ - Slides
+ - Macros
+categories:
+ - Lessons
+ - Macros
+slide-level: 1
+fontsize: 8pt
+header-includes:
+ - \input{../slideOptions.tex}
+
+---
+
+# Welcome to our Data- and Coding-related workshop #
+
+## Purpose: ##
+
+To teach a few tips and tricks for more efficiently managing your
+data, tracking your computer files, understanding appropriate
+analytical approaches, and speeding up the process from code to
+tables.
+
+. . .
+
+## Significance: ##
+
+Topics we cover will help you get more comfortable with data, reduce
+the chance of overlooked errors, and give you more control over your
+work. They are also all important parts of a science movement gaining
+increasing attention -- Reproducible Research.
+
+# Notes and help during this workshop #
+
+Go to this website:
+
+
+
+# What is a macro? #
+
+* SAS has a facility to allow code to be more organized, efficient,
+ and productive for you as the 'coder'
+* Two components:
+
+ - Macro variables
+ - Macros -> A set of commands that can be re-used in different
+ situations to make coding easier.
+
+# What is a macro variable? #
+
+* Method of organizing your code to cut down on typing, reduce errors,
+ and make you more productive
+* Basically act as 'jars' for other variables
+
+. . .
+
+Example code:
+
+ %let jar = BMI FatIntake Activity;
+ proc print data = SASdataset;
+ var &jar; * SAS replaces jar with 'BMI FatIntake Activity';
+ run;
+
+# 4 steps to making a macro #
+
+1. Know what you want the macro to accomplish
+ * Data organization
+ * Statistical analysis
+ * Output printing
+ * Any/all of the above
+2. Type the code you want to run
+ * Data step
+ * Proc step (proc corr, proc glm, proc contents, proc print)
+3. Add macro commands and variables to Step 2
+ * `%macro`
+ * `%mend;` (mend = macro end)
+4. Add macro arguments (basically macro variables)
+5. Save your macros in a separate file
+
+# Not using a macro #
+
+Want to test association between caffeine intake and 3 different
+genetic variants (CYP1A2, ADORA2A, DRD2):
+
+ proc glm data=genes;
+ class CYP1A2 sex smoke;
+ model caff = CYP1A2 BMI sex smoke;
+ lsmeans/ stderr;
+ run;
+
+ proc glm data=genes;
+ class ADORA2A sex smoke;
+ model caff = ADORA2A BMI sex smoke;
+ lsmeans/ stderr;
+ run;
+
+ proc glm data=genes;
+ class DRD2 sex smoke;
+ model caff = DRD2 BMI sex smoke;
+ lsmeans/ stderr;
+ run;
+
+# Why is that undesirable? #
+
+* High risk of making typos or errors and overlooking them
+* Your SAS file can become very long
+* Not ideal for sharing with others, especially once you leave your
+ lab
+
+# Using a macro variable #
+
+Want to test association between caffeine intake and 3 genetic
+variants (CYP1A2, ADORA2A, DRD2):
+
+ %let gene = CYP1A2 ADORA2A DRD2;
+ proc glm data=genes;
+ class &gene sex smoke;
+ model caff = &gene BMI sex smoke;
+ lsmeans/ stderr;
+ run;
+
+. . .
+
+But there is a problem with the above.
+
+# Using a macro #
+
+ %macro glm (data, outcome, predictors, class=);
+ proc glm data=&data ;
+ class &class;
+ model &outcome = &predictors &class;
+ lsmeans/ stderr;
+ run;
+ %mend glm;
+
+ %glm(data = genes, outcome = caff,
+ predictors = CYP1A2);
+
+ /* Or... (for running each gene) */
+ %glm(genes, caff, CYP1A2);
+ %glm(genes, caff, ADORA2A);
+ %glm(genes, caff, DRD2);
+
+# Assignment #
+
+* Make a repetitive code without using a macro. Track this file using Git.
+* Improve this code by making a macro. You can include both codes in the same .sas file. If you want you can even delete the long-style of code and do a "git diff" to see the changes to the file.
+* Give us some "Thoughts" about this workshop as a separate text file. Feel free to include points on how you think you can apply macros in your research.
+* Push both your SAS file and Thoughts file to the appropriate location in the Sandbox repo on GitHub.
+
+# Thanks! #
+
+* Next time: Combining macros with ODS = power!
diff --git a/lessons/ods/assignment.md b/lessons/ods/assignment.md
new file mode 100644
index 0000000..69c3c37
--- /dev/null
+++ b/lessons/ods/assignment.md
@@ -0,0 +1,35 @@
+---
+title: "Assignment: SAS ODS"
+author:
+ - Luke Johnston
+date: 2015-04-13
+fontsize: 12pt
+geometry: margin=1in
+papersize: letterpaper
+layout: page
+sidebar: false
+tag:
+ - Lessons
+ - Assignment
+ - ODS
+categories:
+ - Lessons
+ - ODS
+---
+
+To get better at using ODS, you need to practice it. Try out these
+exercises to get a sense of the power of ODS.
+
+1. Try to create a simple table of some simple statistics (eg. means
+ and standard deviation) *without* using ODS *or* exporting it.
+2. Run a `proc` and determine which ODS object to output.
+3. Extract the ODS object from the `proc`.
+4. Create a simple table from the ODS object *without* customizing it
+ using `data ...; set ...;` *or* exporting it.
+5. Create a simple table from the ODS object, but customize how it
+ looks by using a `data ...; set ...;` *and* export it into a 'csv'
+ format.
+6. Try to incorporate the ODS commands into a macro so that you can
+ quickly analyze and output the results.
+7. Lastly, try to do this on `proc` commands that you normally use in
+ your analysis.
diff --git a/lessons/ods/cheatsheet.md b/lessons/ods/cheatsheet.md
new file mode 100644
index 0000000..5cd84d5
--- /dev/null
+++ b/lessons/ods/cheatsheet.md
@@ -0,0 +1,192 @@
+---
+title: "Cheatsheet: SAS ODS"
+author:
+ - Luke Johnston
+date: 2015-04-12
+fontsize: 12pt
+geometry: margin=1in
+papersize: letterpaper
+layout: page
+sidebar: false
+tag:
+ - Lessons
+ - Cheatsheet
+ - ODS
+categories:
+ - Lessons
+ - ODS
+---
+
+The Output Delivery System (ODS) in SAS allows results from a `proc`
+to be output into a data format so that if you wanted you could
+customize the results in a datastep and eventually output it into a
+file. This is an incredibly useful tool as you can make your analysis
+easier, make it easier to input the results into a manuscript or
+report, and make your research more reproducible and transparent.
+Below are some commands that will help you figure out what to output,
+how to customize the results, and how to save them to a file.
+
+# SAS ODS commands: Some useful or common ones #
+
+## `ods trace on; ... ods trace off;` ##
+
+> When you run a chunk of code with the `ods trace on;` at the start,
+> a `proc` in the middle, and `ods trace off;` at the end, the ODS
+> objects within the `proc` are printed in the log file/window. You
+> can then use that object to output just the object. The name of the
+> ODS object changes depending on what statements are used in the
+> `proc` command. For instance, in `proc anova` if the `lsmeans`
+> statement is provided, `ods trace on;` will show an object that is
+> named `LSMeans`.
+
+> Example code:
+
+ ods trace on;
+ proc means data=sashelp.fish;
+ run;
+ ods trace off;
+
+ /** Comment: The output from this code is: */
+ Output Added:
+ -------------
+ Name: Summary
+ Label: Summary statistics
+ Template: base.summary
+ Path: Means.Summary
+ -------------
+
+## `ods output = ;` ##
+
+> After extracting the name of the ODS object using `ods trace on;`,
+> you can then use the name of the object to output the results
+> specific to the object. For instance, the ODS object name for `proc
+> means` was `Summary`. In order to output the `Summary` object, we
+> would replace it with `` and make our own name up
+> for the output dataset (eg: `meansDS`) in the ``
+> space.
+
+> Example code:
+
+ proc means data=sashelp.fish;
+ ods output Summary = meansDS;
+ run;
+
+ proc print data=meansDS;
+ run;
+
+## `ods listing close; ... ods listing;` ##
+
+> Sometimes when you are outputing an ODS object you also don't want
+> SAS to output the normal results. In this case, you can prevent SAS
+> from sending output to the 'listing' area, either as the `.lst` file
+> or the output window on the SAS editor. That way, you can print
+> only the results that *you* want to see, rather than what *SAS*
+> wants you to see.
+
+> Example code:
+
+ /** Comment: Suppress printing of output */
+ ods listing close;
+ proc means data=sashelp.fish;
+ ods output Summary = meansDS;
+ run;
+ ods listing;
+
+ /** Print the specific output */
+ proc print data=meansDS;
+ run;
+
+## `data ; set ;` ##
+
+> This command should be known to everyone given that, together with
+> all `proc` commands, is the foundation to all SAS commands. This
+> command is included here to reinforce that the `ods output` data is
+> in fact data and can be customized/wrangled. All the commands and
+> code that you use to manage your dataset can also be used to manage
+> your results data (eg. `if .. then ..;`, creating new variables,
+> etc). So for instance, if you only want the probability from an
+> ANOVA test, you can drop all other variables and just output the
+> p-value.
+
+> Example code:
+
+ ods listing close;
+ proc glm data=sashelp.fish;
+ class Species;
+ model Height = Species / ss3;
+ ods output ModelANOVA = modelFish;
+ run;
+ ods listing;
+
+ data modelFish;
+ set modelFish;
+ keep Dependent Source ProbF;
+ run;
+
+ proc print data=modelFish;
+ run;
+
+## `||` ##
+
+> This is the concatenate command. It basically allows you to combine
+> multiple variables and characters together. This is especially
+> useful for modifying ODS output objects so that they can be made
+> into, for example, a table for a report. For instance, if you
+> wanted to make a variable that shows the mean and standard deviation
+> in the form of "mean (SD)", you can use the `||` command to achieve
+> this.
+
+> Example code:
+
+ proc means data=sashelp.fish stackods;
+ ods output Summary = meansDS;
+ run;
+
+ /** Comment: Edit the output results */
+ data meansDS (drop=Mean StdDev);
+ set meansDS;
+ /** In this case, you take the variable 'Mean',
+ combine it using || with a space and a bracket ' ('
+ add the variable StdDev, and lastly add the closing
+ bracket */
+ rawMeanSD = Mean||' ('||
+ StdDev||')';
+ /** If you print the rawMeanSD, there are lots of digits,
+ so you can round it off using `round()` */
+ roundMeanSD = round(Mean, 0.01)||' ('||
+ round(StdDev, 0.01)||')';
+ /** ... However, after rounding, there are extra spaces
+ in the print out. So you can use `strip()` to remove
+ extra white space. */
+ striproundMeanSD = round(Mean, 0.01)||' ('||
+ strip(round(StdDev, 0.01))||')';
+ run;
+
+ proc print data=meansDS;
+ run;
+
+## `proc export ...;` ##
+
+> To make the most of customizing the output of a `proc` into a format
+> that resembles a table, it's probably a good idea to output the
+> results dataset into a file. You can use the `proc export` command
+> to save the dataset into a file of your choice. There are in
+> general four parts to the `proc export` command:
+
+* `data=` is the output dataset.
+* `dbms =` is the output format, eg 'XLS' or 'CSV'. Given the 'csv'
+ format is simply plain text, I would recommend this option.
+* `outfile =` is the name and location of the new output file,
+ eg. 'means.csv'.
+* `replace` tells SAS to overwrite any old file with the same name as
+ the `outfile` name.
+
+> Example code:
+
+ /** Continuing with the dataset from the previous command */
+ proc export data=meansDS
+ dbms = csv
+ outfile = 'means.csv'
+ replace;
+ run;
+
diff --git a/lessons/ods/intro.md b/lessons/ods/intro.md
new file mode 100644
index 0000000..33b09f9
--- /dev/null
+++ b/lessons/ods/intro.md
@@ -0,0 +1,69 @@
+---
+title: "Introduction: SAS ODS"
+author:
+ - Luke Johnston
+date: 2015-04-09
+fontsize: 12pt
+geometry: margin=1in
+papersize: letterpaper
+layout: page
+sidebar: false
+tag:
+ - Lessons
+ - Introduction
+ - ODS
+categories:
+ - Lessons
+ - ODS
+permalink: lessons/ods/
+---
+
+SAS is a system that prints a **lot** of output. Sometimes this can
+be good, however, in general you really only need some of the pieces
+of the output. This is why SAS has another system called Output
+Delivery System, or ODS, to help with this. The power of this becomes
+especially apparent when you have created tables or other output and
+suddenly need to change your analysis (for instance, you were able to
+fill in some of the missing data in your dataset so need to run your
+analysis again). Instead of having to sift through all of the SAS
+output and copy and paste to Excel or other spreadsheet, you can use
+ODS to customize the output *exactly* into the format you need. In
+fact, this technique, known as
+[reproducible research](http://en.wikipedia.org/wiki/Reproducibility#Reproducible_research),
+is increasingly becoming more important and integral to the scientific
+process.
+
+To better appreciate how much time this can save you (especially when
+combined with [SAS macros](../macros/)), ask yourself these questions:
+Has your supervisor or committee ever suggested you change your
+analysis a certain way? Have you ever gotten new data and need to
+update your tables? Has a reviewer ever suggested a revision on your
+manuscript that would require running your analysis again and updating
+your numbers in your manuscript? These are all reasons to learn how
+to use ODS to save you time and reduce your stress.
+
+Our goal for this workshop is to show you how to customize your SAS
+output and convert it into a format that exactly or nearly mimics how
+your table will look like. Depending on the order of the workshops,
+we will also integrate ODS into a macro to save you even *more* time.
+
+# Learning objectives: #
+
+After this workshop, our expectation is that you will be able to:
+
+1. Understand *why* reproducible research is so important and *why*
+ you should be using this technique in *all* your research analyses.
+2. Identify the name of the relevant ODS object from a SAS `proc`.
+3. Extract your desired ODS object.
+4. Customize the data within the ODS object into a format that mimics
+ or is the same as how your final table will look like.
+5. Incorporate an ODS statement into a macro.
+
+# Materials for this lesson: #
+
+* [Slides](slides/)
+* [Cheatsheet](cheatsheet/)
+* [Assignment](assignment/)
+
+Other resources can be found [here](../resources/).
+
diff --git a/lessons/ods/slides.md b/lessons/ods/slides.md
new file mode 100644
index 0000000..e63ab02
--- /dev/null
+++ b/lessons/ods/slides.md
@@ -0,0 +1,102 @@
+---
+title: "From code to tables: Reducing the number of steps in your
+ analysis with SAS ODS"
+author: Luke & Daiva
+date: 2015-04-15
+layout: page
+sidebar: false
+classoption: xcolor=dvipsnames
+tag:
+ - Lessons
+ - Slides
+ - ODS
+categories:
+ - Lessons
+ - ODS
+slide-level: 1
+fontsize: 8pt
+header-includes:
+ - \input{../slideOptions.tex}
+---
+
+# Welcome to our Code As Manuscript workshop! #
+
+## Purpose: ##
+
+To teach a few tips and tricks for more efficiently managing your
+data, tracking your computer files, understanding appropriate
+analytical approaches, and speeding up the process from code to
+tables.
+
+. . .
+
+## Significance: ##
+
+Topics we cover will help you get more comfortable with data, reduce
+the chance of overlooked errors, and give you more control over your
+work. They are also all important parts of a science movement gaining
+increasing attention -- Reproducible Research.
+
+# Caveat: We aren't here to teach statistics or SAS basics #
+
+Need help with stats or SAS? Use these resources!
+
+* U of T Statistical Consulting Services ([click here](http://www.utstat.toronto.edu/wordpress/?page_id=25))
+
+*
+
+*
+
+*
+
+*
+
+# Notes and help during this workshop #
+
+Go to this website:
+
+
+
+Ask questions in the chat, write down notes for others.
+
+Go to our Code As Manuscript website for syllabus, slides, cheatsheet,
+and others:
+
+
+
+# What is ODS? #
+
+* Output Delivery System
+* SAS outputs **A LOT** of results
+* ODS will extract results you are *actually* interested in
+* Cleaner output
+* Easier to re-run analyses with new data and quickly put into
+ manuscript (ie: in tables)
+* Helps with making your research *reproducible*
+* Keeps you saner and less stressed!
+
+# Quick aside: What is reproducible research? #
+
+*
+*
+*
+
+Basically: For any given research project, have the code that can
+easily 'reproduce' the results that are presented in the manuscript.
+
+ODS can help make this more of a reality.
+
+# Identifying the ODS object to use #
+
+ ods trace on;
+ ...
+ ods trace off;
+
+(Switch to live coding)
+
+# Using the ODS object #
+
+ ods output = ;
+
+(Switch to live coding)
+
diff --git a/lessons/r-wrangling/assignment.md b/lessons/r-wrangling/assignment.md
new file mode 100644
index 0000000..dd9f82e
--- /dev/null
+++ b/lessons/r-wrangling/assignment.md
@@ -0,0 +1,56 @@
+---
+title: "Assignment: Data wrangling in R"
+published: true
+author:
+ - Luke W. Johnston
+date: 2015-10
+layout: page
+sidebar: false
+tag:
+ - Lessons
+ - Assignment
+ - Wrangling
+ - R
+categories:
+ - Lessons
+ - Wrangling
+ - R
+output:
+ md_document:
+ variant: markdown_github
+---
+
+## Challenges: Try these out for yourself! ##
+
+Try each of these challenges using only one continuous chain of `%>%` pipes,
+from raw data to final output.
+
+1. Make a new dataframe with the means of Agriculture, Examination, Education, and
+Infant.Mortality for each category of Fertility (hint: convert it into a factor
+by values of >50 vs <50), when Catholic is less than 60 (hint, use `dplyr` commands
+and `gather`). Have the Fertility groups as two columns.
+
+2. Do the same thing as above, but instead make a new dataframe with one column
+with that contains the mean and standard deviation in this format: '00.00 (00.00
+SD)'. Notice that there are two digits after the period.
+
+3. Create a new dataframe with the first column containing the variable names,
+the second column containing which county has the lowest value of the variable,
+and a third column containing the county with the highest value of the variable.
+For example, this is how it should approximately look like:
+
+
+|Variable |Lowest |Highest |
+|:-----------|:--------|:----------|
+|Fertility |Moutier |Courtelary |
+|Agriculture |Delemont |Porrentruy |
+
+### Creating plots (based on the last workshops material)
+
+1. Create a point plot of the means of each variable (not the county). Have the
+variable on the y-axis and the means on the x-axis. As a bonus/option, make the
+graph prettier.
+
+2. Expand on challenge 4, but split the means up by fertility (like in challenge
+1). The graph should have two dots for each variable representing the means for
+each group of fertility.
diff --git a/lessons/r-wrangling/cheatsheet.md b/lessons/r-wrangling/cheatsheet.md
new file mode 100644
index 0000000..ccad661
--- /dev/null
+++ b/lessons/r-wrangling/cheatsheet.md
@@ -0,0 +1,323 @@
+---
+title: "Cheatsheet: Data wrangling in R"
+published: true
+author:
+ - Luke W. Johnston
+date: 2015-01
+layout: page
+sidebar: false
+tag:
+ - Lessons
+ - Cheatsheet
+ - Wrangling
+ - R
+categories:
+ - Lessons
+ - Wrangling
+ - R
+output:
+ md_document:
+ variant: markdown_github
+ toc: true
+---
+
+
+
+R is a statistical computing environment to analyze data and write programs.
+Getting the data into an analyzable form is the hardest part of an analysis.
+Here is a cheatsheet of useful and/or common commands. For a more detailed
+cheatsheet, [check out the resource page](../resources/).
+
+# R data wrangling commands: Some useful or common ones #
+
+## `write.csv` or `write.table` ##
+
+> Save the R object or dataframe (aka. dataset) in a `.csv` file (comma
+separated values). Using `write.table`, you can export a number of different
+file formats.
+
+> Example code:
+
+
+{% highlight r %}
+## Export
+write.csv(swiss, file = 'swiss.csv')
+
+## Which is the same as:
+write.table(swiss, file = 'swiss.csv', sep = ',')
+{% endhighlight %}
+
+## `read.csv` or `read.table` ##
+
+> Similar to `write.table`. Imports a file (eg. `csv`).
+
+> Example code:
+
+
+{% highlight r %}
+write.csv(swiss, file = 'swiss.csv')
+read.csv('swiss.csv')
+{% endhighlight %}
+
+## `head`, `names`, `str`, `summary` ##
+
+> These four commands give you a brief overview of your dataframe or R object.
+`head` lets you see the first 6 rows. `names` lets you see the variable names in
+the dataframe. `str` lets you see the structure of your R object/dataframe,
+showing you what variables there are, and what the type is (eg. number, integer,
+character, etc). `summary` is very useful to get a quick overview of the mean,
+median, frequency, and other basic statistics of each variable in the dataframe.
+
+> Example code:
+
+
+{% highlight r %}
+head(swiss)
+names(swiss)
+str(swiss)
+summary(swiss)
+class(swiss)
+{% endhighlight %}
+
+## `%>%` ##
+
+> The pipe function is a general purpose, extremely powerful tool for making
+your code easier to read and quicker to type out. It takes the output from the
+left hand side and puts it into the right hand command. The `.` command tells
+the pipe to use the output from the left hand side, which you sometimes have to
+do for some functions/commands (like `lm()`).
+
+> Example code:
+
+
+{% highlight r %}
+library(dplyr)
+## This is the package that the pipe comes from
+library(magrittr)
+
+## These are the same
+sum(1:10)
+1:10 %>% sum
+
+## These all do the same thing
+head(swiss)
+swiss %>% head
+swiss %>% head()
+swiss %>% head(.)
+{% endhighlight %}
+
+## `tbl_df` ##
+
+> Sets the `tbl` attribute to the dataframe, which makes the printing of the
+dataframe prettier.
+
+> Example code:
+
+
+{% highlight r %}
+library(dplyr)
+## These are the same
+tbl_df(ds)
+ds %>% tbl_df
+ds %>% tbl_df()
+ds %>% tbl_df(.)
+{% endhighlight %}
+
+## `select` ##
+
+> Get only the columns/variables you want from a dataframe. You can select
+variables based on pattern or if it contains some letter.
+
+> Example code:
+
+
+{% highlight r %}
+library(dplyr)
+## These are the same
+select(swiss, Education, Catholic, Fertility)
+swiss %>% select(Education, Catholic, Fertility)
+swiss %>% select(., Education, Catholic, Fertility)
+
+## Exclude variable(s)
+swiss %>% select(-Education, -Catholic)
+
+## Select variables based on name or pattern
+swiss %>% select(starts_with('E'), contains('Fert'), matches('mort'))
+{% endhighlight %}
+
+## `rename` ##
+
+> Change the name of a variable/column. The new name is on the left hand side,
+so `newname = oldname`. You can also rename using the `select` command, but it
+will only select what variables you give it, while rename keeps the dataframe as
+is, as it only renames.
+
+> Example code:
+
+
+{% highlight r %}
+library(dplyr)
+## These are the same
+rename(swiss, edu = Education)
+swiss %>% rename(edu = Education)
+
+## Multiple renaming
+swiss %>% rename(edu = Education, fert = Fertility)
+
+## If you want to use select, but get the same functionality as rename, use the
+## everything() function to select all other variables in the dataframe
+swiss %>% select(edu = Education, everything())
+{% endhighlight %}
+
+## `filter` ##
+
+> Subset a dataframe based on a condition of a variable. Filtering splits the
+dataframe by the value of the row. Common conditions include `<` less than, `>`
+greater than, `==` equals, `>=` or `<=` greater/less than or equal to.
+
+> Example code:
+
+
+{% highlight r %}
+library(dplyr)
+## These are the same
+filter(swiss, Catholic < 20, Examination == 15)
+swiss %>% filter(Catholic < 20, Examination == 15)
+swiss %>% filter(., Catholic < 20, Examination == 15)
+
+## For string/factor variables
+swiss %>% filter(X == 'Aigle')
+{% endhighlight %}
+
+## `mutate` ##
+
+> Create a new column. A command that may be used is the `ifelse()` function
+that assigns a value based on the condition.
+
+> Example code:
+
+
+{% highlight r %}
+library(dplyr)
+## These are the same
+mutate(swiss, Infertile = ifelse(Fertility < 50, 'yes', 'no'))
+swiss %>% mutate(Infertile = ifelse(Fertility < 50, 'yes', 'no'))
+swiss %>% mutate(., Infertile = ifelse(Fertility < 50, 'yes', 'no'))
+
+## Or..
+swiss %>% mutate(Test = 'yes', Number = 10)
+{% endhighlight %}
+
+## `arrange` ##
+
+> Sort/order a dataframe by a variable. Sort by the order of the variables
+given (eg. `arrange(var1, var2)` sorts first by `var1` than by `var2`).
+
+> Example code:
+
+
+{% highlight r %}
+library(dplyr)
+## These are the same
+arrange(swiss, Education, Examination)
+swiss %>% arrange(Education, Examination)
+swiss %>% arrange(., Education, Examination)
+
+## Or to do it descending
+swiss %>% arrange(desc(Education))
+{% endhighlight %}
+
+## `group_by` ##
+
+> Assigns the attribute `grouped_df` to the dataframe, which on it's own does
+nothing but when used in conjunction with `summarise` or `mutate` does the
+following commands based on the grouping.
+
+> Example code:
+
+
+{% highlight r %}
+library(dplyr)
+swiss %>%
+ mutate(EarlyDeath = ifelse(Infant.Mortality >= 50, 'yes', 'no')) %>%
+ group_by(EarlyDeath)
+{% endhighlight %}
+
+## `summarise` ##
+
+> Create a new column of values, usually using a descriptive statistic function
+such as `mean()` or `median()`, as well as informational functions like `n()`
+for sample size. This function is best used with `group_by()`.
+
+> Example code:
+
+
+{% highlight r %}
+library(dplyr)
+swiss %>%
+ mutate(Educated = ifelse(Education >= 50, 'yes', 'no')) %>%
+ group_by(Educated) %>%
+ str()
+ summarise(mean = mean(Agriculture))
+{% endhighlight %}
+
+
+## `gather` ##
+
+> Convert a wide dataframe to a long dataframe. This creates a dataframe with
+at least two new variables, one containing the names of the original variables
+and the other containing the values of the variables. You can include or
+exclude certain variables by selecting the variable you want to include or
+exclude (with a `-`) after the name of the two new variables.
+
+> Example code:
+
+
+{% highlight r %}
+library(dplyr)
+library(tidyr)
+## These are the same
+gather(swiss, Measure, Value)
+swiss %>% gather(Measure, Value)
+swiss %>% gather(., Measure, Value)
+
+## Or exclude certain variables
+swiss %>% add_rownames() %>% gather(Measure, Value, -rowname)
+
+## Or include only some variables
+swiss %>% gather(Measure, Value, Education, Fertility, Infant.Mortality)
+{% endhighlight %}
+
+## `spread` ##
+
+> Does the opposite of `gather` by converting long dataframes to wide
+dataframes.
+
+> Example code:
+
+
+{% highlight r %}
+library(dplyr)
+library(tidyr)
+swiss %>%
+ add_rownames() %>%
+ gather(Measure, Value, -rowname) %>%
+ spread(Measure, Value)
+{% endhighlight %}
+
+# Combined example using (almost) all functions:
+
+
+{% highlight r %}
+swiss %>%
+ add_rownames() %>%
+ tbl_df() %>%
+ filter(Infant.Mortality >= 15) %>%
+ mutate(Religious = ifelse(Catholic <= 50, 'no', 'yes')) %>%
+ select(Education, Agriculture, Examination, Religious) %>%
+ gather(Measure, Value, -Religious) %>%
+ group_by(Measure, Religious) %>%
+ summarise(mean = mean(Value)) %>%
+ spread(Measure, mean)
+{% endhighlight %}
+
diff --git a/lessons/r-wrangling/intro.md b/lessons/r-wrangling/intro.md
new file mode 100644
index 0000000..6d88165
--- /dev/null
+++ b/lessons/r-wrangling/intro.md
@@ -0,0 +1,764 @@
+---
+title: "Introduction: Data wrangling in R"
+published: true
+author:
+ - Luke W. Johnston
+date: 2015-10
+layout: page
+sidebar: false
+tag:
+ - Lessons
+ - Introduction
+ - Wrangling
+ - R
+categories:
+ - Lessons
+ - Wrangling
+ - R
+permalink: lessons/r-wrangling/
+output:
+ md_document:
+ variant: markdown_github
+ toc: true
+---
+
+R was developed by statisticians to do statistical work. As such, embedded
+within R are capabilities to easily wrangle and manage data, to have data in a
+format that can be used for further data analysis, and to work data set type
+objects (called dataframes in R). There are also excellent packages available
+to make data wrangling much easier in R. These packages are `dplyr` and `tidyr`
+packages. A minor assumption: the data you are importing/using is fairly clean
+(as in, no large amount of missing values, no data entry errors or fixes needed,
+etc).
+
+# Learning objectives:
+
+1. How to import/export your data
+2. How to view the structure of your data
+3. How to wrangle data into an analyzable format
+
+# Materials for this lesson:
+
+- [Slides](slides/)
+- [Cheatsheet](cheatsheet/)
+- [Assignment](assignment/)
+
+Other resources can be found [here](../resources/).
+
+# Let's get wrangling, the basics
+
+## Import/export your data
+
+You'll need to import your data into R to analyze it. We'll assume the data is
+already cleaned and ready for analysis. If at any time you need help with a
+command, use the `?` command along with the command you need help with (eg.
+`?write.csv`). R comes with many internal datasets that you can practice on.
+The one I'm going to use is the `swiss` dataset.
+
+
+{% highlight r %}
+## Export/save to file
+write.csv(swiss, file = 'swiss.csv')
+## Import/read from file
+ds <- read.csv('swiss.csv')
+{% endhighlight %}
+
+## Viewing your data
+
+R has several very useful tools for quickly viewing your data. `head()` shows
+the first few rows of a dataframe (a structure for storing data that can include
+numbers, integers, factors, strings, etc). `names()` shows the column names.
+`str()` shows the structure, such as what the object is, and its contents (ie.
+column names and types). `summary()` shows a quick description of the summary
+statistics (means, median, frequency) for each of your columns.
+
+
+{% highlight r %}
+head(ds)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## X Fertility Agriculture Examination Education Catholic
+## 1 Courtelary 80.2 17.0 15 12 9.96
+## 2 Delemont 83.1 45.1 6 9 84.84
+## 3 Franches-Mnt 92.5 39.7 5 5 93.40
+## 4 Moutier 85.8 36.5 12 7 33.77
+## 5 Neuveville 76.9 43.5 17 15 5.16
+## 6 Porrentruy 76.1 35.3 9 7 90.57
+## Infant.Mortality
+## 1 22.2
+## 2 22.2
+## 3 20.2
+## 4 20.3
+## 5 20.6
+## 6 26.6
+{% endhighlight %}
+
+
+
+{% highlight r %}
+names(ds)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] "X" "Fertility" "Agriculture"
+## [4] "Examination" "Education" "Catholic"
+## [7] "Infant.Mortality"
+{% endhighlight %}
+
+
+
+{% highlight r %}
+str(ds)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## 'data.frame': 47 obs. of 7 variables:
+## $ X : Factor w/ 47 levels "Aigle","Aubonne",..: 8 9 12 26 28 34 5 13 15 38 ...
+## $ Fertility : num 80.2 83.1 92.5 85.8 76.9 76.1 83.8 92.4 82.4 82.9 ...
+## $ Agriculture : num 17 45.1 39.7 36.5 43.5 35.3 70.2 67.8 53.3 45.2 ...
+## $ Examination : int 15 6 5 12 17 9 16 14 12 16 ...
+## $ Education : int 12 9 5 7 15 7 7 8 7 13 ...
+## $ Catholic : num 9.96 84.84 93.4 33.77 5.16 ...
+## $ Infant.Mortality: num 22.2 22.2 20.2 20.3 20.6 26.6 23.6 24.9 21 24.4 ...
+{% endhighlight %}
+
+
+
+{% highlight r %}
+summary(ds)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## X Fertility Agriculture Examination
+## Aigle : 1 Min. :35.00 Min. : 1.20 Min. : 3.00
+## Aubonne : 1 1st Qu.:64.70 1st Qu.:35.90 1st Qu.:12.00
+## Avenches: 1 Median :70.40 Median :54.10 Median :16.00
+## Boudry : 1 Mean :70.14 Mean :50.66 Mean :16.49
+## Broye : 1 3rd Qu.:78.45 3rd Qu.:67.65 3rd Qu.:22.00
+## Conthey : 1 Max. :92.50 Max. :89.70 Max. :37.00
+## (Other) :41
+## Education Catholic Infant.Mortality
+## Min. : 1.00 Min. : 2.150 Min. :10.80
+## 1st Qu.: 6.00 1st Qu.: 5.195 1st Qu.:18.15
+## Median : 8.00 Median : 15.140 Median :20.00
+## Mean :10.98 Mean : 41.144 Mean :19.94
+## 3rd Qu.:12.00 3rd Qu.: 93.125 3rd Qu.:21.70
+## Max. :53.00 Max. :100.000 Max. :26.60
+##
+{% endhighlight %}
+
+# Wrangling your data, `dplyr` style
+
+Data wrangling can be a bit tedious in base R (R without packages), so we'll be
+using two packages designed to make this easier. Load the `tidyr` and `dplyr`
+packages by using the `library()` function. `dplyr` comes with a `%>%` pipe
+function (via the `magrittr` package), which works similar to how the Bash shell
+`|` pipe works (for those familiar with Bash, ie. those who use Mac or Linux).
+The command on the right-hand side takes the output from the command on the
+left-hand side, just like how a plumbing pipe works for water. `tbl_df` makes
+the object into a `tbl` class, making printing of the output nicer.
+
+
+{% highlight r %}
+library(dplyr)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+##
+## Attaching package: 'dplyr'
+##
+## The following objects are masked from 'package:stats':
+##
+## filter, lag
+##
+## The following objects are masked from 'package:base':
+##
+## intersect, setdiff, setequal, union
+{% endhighlight %}
+
+
+
+{% highlight r %}
+library(tidyr)
+
+## Compare
+head(ds)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## X Fertility Agriculture Examination Education Catholic
+## 1 Courtelary 80.2 17.0 15 12 9.96
+## 2 Delemont 83.1 45.1 6 9 84.84
+## 3 Franches-Mnt 92.5 39.7 5 5 93.40
+## 4 Moutier 85.8 36.5 12 7 33.77
+## 5 Neuveville 76.9 43.5 17 15 5.16
+## 6 Porrentruy 76.1 35.3 9 7 90.57
+## Infant.Mortality
+## 1 22.2
+## 2 22.2
+## 3 20.2
+## 4 20.3
+## 5 20.6
+## 6 26.6
+{% endhighlight %}
+
+
+
+{% highlight r %}
+## With:
+tbl_df(ds)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## Source: local data frame [47 x 7]
+##
+## X Fertility Agriculture Examination Education Catholic
+## (fctr) (dbl) (dbl) (int) (int) (dbl)
+## 1 Courtelary 80.2 17.0 15 12 9.96
+## 2 Delemont 83.1 45.1 6 9 84.84
+## 3 Franches-Mnt 92.5 39.7 5 5 93.40
+## 4 Moutier 85.8 36.5 12 7 33.77
+## 5 Neuveville 76.9 43.5 17 15 5.16
+## 6 Porrentruy 76.1 35.3 9 7 90.57
+## 7 Broye 83.8 70.2 16 7 92.85
+## 8 Glane 92.4 67.8 14 8 97.16
+## 9 Gruyere 82.4 53.3 12 7 97.67
+## 10 Sarine 82.9 45.2 16 13 91.38
+## .. ... ... ... ... ... ...
+## Variables not shown: Infant.Mortality (dbl)
+{% endhighlight %}
+
+
+
+{% highlight r %}
+## Now put the tbl dataset into a new object
+ds2 <- tbl_df(ds)
+ds2
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## Source: local data frame [47 x 7]
+##
+## X Fertility Agriculture Examination Education Catholic
+## (fctr) (dbl) (dbl) (int) (int) (dbl)
+## 1 Courtelary 80.2 17.0 15 12 9.96
+## 2 Delemont 83.1 45.1 6 9 84.84
+## 3 Franches-Mnt 92.5 39.7 5 5 93.40
+## 4 Moutier 85.8 36.5 12 7 33.77
+## 5 Neuveville 76.9 43.5 17 15 5.16
+## 6 Porrentruy 76.1 35.3 9 7 90.57
+## 7 Broye 83.8 70.2 16 7 92.85
+## 8 Glane 92.4 67.8 14 8 97.16
+## 9 Gruyere 82.4 53.3 12 7 97.67
+## 10 Sarine 82.9 45.2 16 13 91.38
+## .. ... ... ... ... ... ...
+## Variables not shown: Infant.Mortality (dbl)
+{% endhighlight %}
+
+## Select columns
+
+Often times, you want to select only some of the columns or variables from a
+dataset. For that we use the `select` command, which does as it says. Note the
+use of the `%>%` operator. This allows you to chain commands together, letting
+you do more with only a few commands.
+
+
+{% highlight r %}
+ds2 %>%
+ select(Education, Catholic, Fertility)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## Source: local data frame [47 x 3]
+##
+## Education Catholic Fertility
+## (int) (dbl) (dbl)
+## 1 12 9.96 80.2
+## 2 9 84.84 83.1
+## 3 5 93.40 92.5
+## 4 7 33.77 85.8
+## 5 15 5.16 76.9
+## 6 7 90.57 76.1
+## 7 7 92.85 83.8
+## 8 8 97.16 92.4
+## 9 7 97.67 82.4
+## 10 13 91.38 82.9
+## .. ... ... ...
+{% endhighlight %}
+
+The real power with using the `select()` function comes when you combine it with
+[regular expressions (regexp)](http://www.regular-expressions.info/), or rather pattern
+searching. `dplyr` has several pattern searching functions, including
+`starts_with()`, `contains()`, and the most powerful `matches()`. The function
+`matches()` uses regexp, which are special commands that use
+certain, unique syntax for searching for patterns. For example, `^string` means
+that `string` is the first character, `string$` means that `string` is last
+character, `string|strung` searches for either `string` or `strung`, etc.
+regexp syntax are nearly a language to themselves, so use
+[StackOverflow](http://stackoverflow.com/) and Google as much as you can!
+
+Ok, so lets say you want to search for variables that have certain patterns:
+
+
+{% highlight r %}
+ds2 %>%
+ select(contains('Edu'), starts_with('Cath'))
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## Source: local data frame [47 x 2]
+##
+## Education Catholic
+## (int) (dbl)
+## 1 12 9.96
+## 2 9 84.84
+## 3 5 93.40
+## 4 7 33.77
+## 5 15 5.16
+## 6 7 90.57
+## 7 7 92.85
+## 8 8 97.16
+## 9 7 97.67
+## 10 13 91.38
+## .. ... ...
+{% endhighlight %}
+
+
+
+{% highlight r %}
+## Or more simplified
+ds2 %>%
+ select(matches('Edu|Cath'))
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## Source: local data frame [47 x 2]
+##
+## Education Catholic
+## (int) (dbl)
+## 1 12 9.96
+## 2 9 84.84
+## 3 5 93.40
+## 4 7 33.77
+## 5 15 5.16
+## 6 7 90.57
+## 7 7 92.85
+## 8 8 97.16
+## 9 7 97.67
+## 10 13 91.38
+## .. ... ...
+{% endhighlight %}
+
+You can see that if you have many variables that have a common structure to
+their name, you can quickly select all those variables by using functions such
+as `matches()`.
+
+## Rename columns
+
+You can rename columns using the `rename` command (the new name is on the left
+hand side, so `newname = oldname`).
+
+
+{% highlight r %}
+ds2 %>%
+ rename(County = X)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## Source: local data frame [47 x 7]
+##
+## County Fertility Agriculture Examination Education Catholic
+## (fctr) (dbl) (dbl) (int) (int) (dbl)
+## 1 Courtelary 80.2 17.0 15 12 9.96
+## 2 Delemont 83.1 45.1 6 9 84.84
+## 3 Franches-Mnt 92.5 39.7 5 5 93.40
+## 4 Moutier 85.8 36.5 12 7 33.77
+## 5 Neuveville 76.9 43.5 17 15 5.16
+## 6 Porrentruy 76.1 35.3 9 7 90.57
+## 7 Broye 83.8 70.2 16 7 92.85
+## 8 Glane 92.4 67.8 14 8 97.16
+## 9 Gruyere 82.4 53.3 12 7 97.67
+## 10 Sarine 82.9 45.2 16 13 91.38
+## .. ... ... ... ... ... ...
+## Variables not shown: Infant.Mortality (dbl)
+{% endhighlight %}
+
+## Filter rows
+
+Another common task in data wrangling is subsetting your dataset. You can
+subset the dataset using `filter`. Note the double equal sign `==` for testing
+if 'Examination' is equal to 15. A single `=` is used for something else
+(assigning things to objects or using them in functions/commands).
+
+
+{% highlight r %}
+## For continuous/number data
+ds2 %>%
+ filter(Catholic < 20, Examination == 15)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## Source: local data frame [3 x 7]
+##
+## X Fertility Agriculture Examination Education Catholic
+## (fctr) (dbl) (dbl) (int) (int) (dbl)
+## 1 Courtelary 80.2 17.0 15 12 9.96
+## 2 Yverdon 65.4 49.5 15 8 6.10
+## 3 Val de Ruz 77.6 37.6 15 7 4.97
+## Variables not shown: Infant.Mortality (dbl)
+{% endhighlight %}
+
+
+
+{% highlight r %}
+## Or for 'string' (words or letters) data
+ds2 %>%
+ filter(X == 'Aigle')
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## Source: local data frame [1 x 7]
+##
+## X Fertility Agriculture Examination Education Catholic
+## (fctr) (dbl) (dbl) (int) (int) (dbl)
+## 1 Aigle 64.1 62 21 12 8.52
+## Variables not shown: Infant.Mortality (dbl)
+{% endhighlight %}
+
+## Create new columns or clean up existing ones
+
+If you want to create a new column, you use the `mutate` command. The
+`ifelse()` command lets you use a condition to have different values depending
+on the condition.
+
+
+{% highlight r %}
+ds2 %>%
+ mutate(Testing = 'yes',
+ Infertile = ifelse(Fertility < 50, 'yes', 'no'))
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## Source: local data frame [47 x 9]
+##
+## X Fertility Agriculture Examination Education Catholic
+## (fctr) (dbl) (dbl) (int) (int) (dbl)
+## 1 Courtelary 80.2 17.0 15 12 9.96
+## 2 Delemont 83.1 45.1 6 9 84.84
+## 3 Franches-Mnt 92.5 39.7 5 5 93.40
+## 4 Moutier 85.8 36.5 12 7 33.77
+## 5 Neuveville 76.9 43.5 17 15 5.16
+## 6 Porrentruy 76.1 35.3 9 7 90.57
+## 7 Broye 83.8 70.2 16 7 92.85
+## 8 Glane 92.4 67.8 14 8 97.16
+## 9 Gruyere 82.4 53.3 12 7 97.67
+## 10 Sarine 82.9 45.2 16 13 91.38
+## .. ... ... ... ... ... ...
+## Variables not shown: Infant.Mortality (dbl), Testing (chr), Infertile
+## (chr)
+{% endhighlight %}
+
+However, it's fairly common that you need to do some data janitorial work by
+cleaning up an existing column. For example, in a dataset with a 'Sex' variable,
+some values had data entry errors in spelling, such as 'fmale' when it should be
+'female'. This needs to be fixed and can be done fairly easily in R. So let's
+'pretend that all words starting with 'G' in the `X` (county) column should
+'actually be 'J' and that all words with an 'e' at the end should be removed.
+'For this, we will use the `gsub()` command within `mutate()`, which will
+'*g*lobablly *sub*stitute a pattern with the replacement.
+
+
+{% highlight r %}
+ds2 %>%
+ mutate(
+ X = gsub(pattern = '^G', replacement = 'J', X),
+ X = gsub(pattern = 'e$', replacement = '', X)
+ )
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## Source: local data frame [47 x 7]
+##
+## X Fertility Agriculture Examination Education Catholic
+## (chr) (dbl) (dbl) (int) (int) (dbl)
+## 1 Courtelary 80.2 17.0 15 12 9.96
+## 2 Delemont 83.1 45.1 6 9 84.84
+## 3 Franches-Mnt 92.5 39.7 5 5 93.40
+## 4 Moutier 85.8 36.5 12 7 33.77
+## 5 Neuvevill 76.9 43.5 17 15 5.16
+## 6 Porrentruy 76.1 35.3 9 7 90.57
+## 7 Broy 83.8 70.2 16 7 92.85
+## 8 Jlan 92.4 67.8 14 8 97.16
+## 9 Jruyer 82.4 53.3 12 7 97.67
+## 10 Sarin 82.9 45.2 16 13 91.38
+## .. ... ... ... ... ... ...
+## Variables not shown: Infant.Mortality (dbl)
+{% endhighlight %}
+
+Notice the `^` and `$` characters. Those are special syntax symbols used in
+regexp commands. We introduced them above, but we'll quickly go over
+it again. These special symbols perform certain functions. In this case `^G`
+means for all "G" that are at the start of the string/character, while `e$`
+means for all "e" that are at the end of a string. Or let"s say that all "mont",
+"mout", and "mnt" should actually be "ment". We can do some cleaning fairly easily
+here.
+
+
+{% highlight r %}
+ds2 %>%
+ mutate(X = gsub('mont|mnt|mout', 'ment', X, ignore.case = TRUE))
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## Source: local data frame [47 x 7]
+##
+## X Fertility Agriculture Examination Education Catholic
+## (chr) (dbl) (dbl) (int) (int) (dbl)
+## 1 Courtelary 80.2 17.0 15 12 9.96
+## 2 Delement 83.1 45.1 6 9 84.84
+## 3 Franches-ment 92.5 39.7 5 5 93.40
+## 4 mentier 85.8 36.5 12 7 33.77
+## 5 Neuveville 76.9 43.5 17 15 5.16
+## 6 Porrentruy 76.1 35.3 9 7 90.57
+## 7 Broye 83.8 70.2 16 7 92.85
+## 8 Glane 92.4 67.8 14 8 97.16
+## 9 Gruyere 82.4 53.3 12 7 97.67
+## 10 Sarine 82.9 45.2 16 13 91.38
+## .. ... ... ... ... ... ...
+## Variables not shown: Infant.Mortality (dbl)
+{% endhighlight %}
+
+Regular expressions are incredibly powerful, but also can be confusing. Make
+sure to check out [our resources page](/lessons/resources/) for links to sites
+that explain regexp in more detail.
+
+## Chaining pipes
+
+We can start chaining these commands together using the `%>%` pipe command.
+There is no limit to how long a chain can be. Chaining commands together using
+the pipe command makes your code easier to read, makes you type out your code
+faster, and makes it easier for you to go from thinking of an analysis to
+actually conducting it. `arrange` sorts/orders/re-arranges the column Education
+in ascending order.
+
+
+{% highlight r %}
+ds2 %>%
+ filter(Catholic > 20) %>%
+ select(County = X, ## This renames the variable, just like the rename() command
+ Education, Fertility, Agriculture) %>%
+ arrange(Education) %>%
+ mutate(infertile = ifelse(Fertility < 50, 'yes', 'no'))
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## Source: local data frame [21 x 5]
+##
+## County Education Fertility Agriculture infertile
+## (fctr) (int) (dbl) (dbl) (chr)
+## 1 Echallens 2 68.3 72.6 no
+## 2 Conthey 2 75.5 85.9 no
+## 3 Herens 2 77.3 89.7 no
+## 4 Monthey 3 79.4 64.9 no
+## 5 Sierre 3 92.2 84.6 no
+## 6 Franches-Mnt 5 92.5 39.7 no
+## 7 Veveyse 6 87.1 64.5 no
+## 8 Entremont 6 69.3 84.9 no
+## 9 Martigwy 6 70.5 78.2 no
+## 10 Moutier 7 85.8 36.5 no
+## .. ... ... ... ... ...
+{% endhighlight %}
+
+## Re-organize your data (using `tidyr`)
+
+To get the data into a nicer and more analyable format, you can use the `tidyr`
+package. See what `gather` does in the code below. Then see what `spread`
+does. Note that you can remove a column by having a minus `-` sign in front of
+a variable when you use `select`.
+
+
+{% highlight r %}
+## Compare this:
+ds2 %>%
+ select(-Infant.Mortality) %>%
+ rename(County = X)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## Source: local data frame [47 x 6]
+##
+## County Fertility Agriculture Examination Education Catholic
+## (fctr) (dbl) (dbl) (int) (int) (dbl)
+## 1 Courtelary 80.2 17.0 15 12 9.96
+## 2 Delemont 83.1 45.1 6 9 84.84
+## 3 Franches-Mnt 92.5 39.7 5 5 93.40
+## 4 Moutier 85.8 36.5 12 7 33.77
+## 5 Neuveville 76.9 43.5 17 15 5.16
+## 6 Porrentruy 76.1 35.3 9 7 90.57
+## 7 Broye 83.8 70.2 16 7 92.85
+## 8 Glane 92.4 67.8 14 8 97.16
+## 9 Gruyere 82.4 53.3 12 7 97.67
+## 10 Sarine 82.9 45.2 16 13 91.38
+## .. ... ... ... ... ... ...
+{% endhighlight %}
+
+
+
+{% highlight r %}
+## With this:
+ds2 %>%
+ select(-Infant.Mortality) %>%
+ rename(County = X) %>%
+ gather(Measure, Value, -County)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## Source: local data frame [235 x 3]
+##
+## County Measure Value
+## (fctr) (fctr) (dbl)
+## 1 Courtelary Fertility 80.2
+## 2 Delemont Fertility 83.1
+## 3 Franches-Mnt Fertility 92.5
+## 4 Moutier Fertility 85.8
+## 5 Neuveville Fertility 76.9
+## 6 Porrentruy Fertility 76.1
+## 7 Broye Fertility 83.8
+## 8 Glane Fertility 92.4
+## 9 Gruyere Fertility 82.4
+## 10 Sarine Fertility 82.9
+## .. ... ... ...
+{% endhighlight %}
+
+
+
+{% highlight r %}
+## And back again:
+ds2 %>%
+ select(-Infant.Mortality) %>%
+ rename(County = X) %>%
+ gather(Measure, Value, -County) %>%
+ spread(Measure, Value)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## Source: local data frame [47 x 6]
+##
+## County Fertility Agriculture Examination Education Catholic
+## (fctr) (dbl) (dbl) (dbl) (dbl) (dbl)
+## 1 Aigle 64.1 62.0 21 12 8.52
+## 2 Aubonne 66.9 67.5 14 7 2.27
+## 3 Avenches 68.9 60.7 19 12 4.43
+## 4 Boudry 70.4 38.4 26 12 5.62
+## 5 Broye 83.8 70.2 16 7 92.85
+## 6 Conthey 75.5 85.9 3 2 99.71
+## 7 Cossonay 61.7 69.3 22 5 2.82
+## 8 Courtelary 80.2 17.0 15 12 9.96
+## 9 Delemont 83.1 45.1 6 9 84.84
+## 10 Echallens 68.3 72.6 18 2 24.20
+## .. ... ... ... ... ... ...
+{% endhighlight %}
+
+## Summarise variables
+
+Combined with `dplyr`'s `group_by` and `summarise` you can quickly summarise
+data or do further, more complicated analyses. `group_by` makes it so further
+analyses or operations work on the groups. `summarise` transforms the data to
+only contain the new variable(s) created, in this case the mean, as well as the
+grouping variable.
+
+
+{% highlight r %}
+ds2 %>%
+ select(-X) %>%
+ gather(Measure, Value) %>%
+ group_by(Measure) %>%
+ summarise(mean = mean(Value),
+ sampleSize = n())
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## Source: local data frame [6 x 3]
+##
+## Measure mean sampleSize
+## (fctr) (dbl) (int)
+## 1 Fertility 70.14255 47
+## 2 Agriculture 50.65957 47
+## 3 Examination 16.48936 47
+## 4 Education 10.97872 47
+## 5 Catholic 41.14383 47
+## 6 Infant.Mortality 19.94255 47
+{% endhighlight %}
+
+## Other useful and powerful examples
+
+You can do some really powerful things with `dplyr` and `tidyr` functions. For
+example, you can run each combination of independent and dependent variables in
+a linear regression (`lm()`) using `gather()` and the `dplyr` `do()` command,
+rather than running each individually. To make the output from the `lm()`
+easier to combine and understand, we use the `tidy()` command from the `broom`
+package (`::` tells R we want to use the `tidy()` function from the `broom`
+package). If you want more details on how to use this set up,
+[check out my blog post about it](http://www.lukewjohnston.com/blog/loops-forests-multiple-linear-regressions/).
+
+
+{% highlight r %}
+ds2 %>%
+ select(-X) %>%
+ gather(Indep, Xvalue, Fertility, Agriculture) %>%
+ gather(Dep, Yvalue, Education, Catholic) %>%
+ group_by(Dep, Indep) %>%
+ do(lm(Yvalue ~ Xvalue + Infant.Mortality + Examination, data = .) %>%
+ broom::tidy())
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## Error in tidy.lm(.): could not find function "is"
+{% endhighlight %}
diff --git a/lessons/r-wrangling/slides.md b/lessons/r-wrangling/slides.md
new file mode 100644
index 0000000..7657f66
--- /dev/null
+++ b/lessons/r-wrangling/slides.md
@@ -0,0 +1,32 @@
+---
+title: "Data wrangling in R"
+published: true
+author: Luke W. Johnston
+date: 2015-10
+layout: page
+sidebar: false
+tag:
+ - Lessons
+ - Slides
+ - Wrangling
+ - R
+categories:
+ - Lessons
+ - Wrangling
+ - R
+output: slidy_presentation
+---
+
+## Learning expectations ##
+
+- How to import/export your data
+- How to view the structure of your data
+- How to wrangle data into an analyzable format
+
+## 4 main concepts: ##
+
+- **Getting the data**: read.table, write.table
+- **View the data**: str, summary, names, head
+- **Working the data (dplyr):** (tbl\_df), select, filter, mutate,
+ summarise, arrange, rename, group\_by, `%>%` pipe
+- **(Re)Organize the data (tidyr):** gather, spread
diff --git a/lessons/resources.md b/lessons/resources.md
new file mode 100644
index 0000000..477043f
--- /dev/null
+++ b/lessons/resources.md
@@ -0,0 +1,114 @@
+---
+title: Resources
+author:
+ - Luke Johnston
+date: 2015-04-12
+layout: page
+sidebar: false
+tag:
+ - Resources
+categories:
+ - Resources
+---
+
+# Content: #
+
+* [Reproductibility](#reproducibility)
+* [Git](#git)
+* [SAS ODS](#sas-ods)
+
+# Reproducibility #
+
+* Wilson G, Aruliah DA, Brown CT, Chue Hong NP, Davis M, et
+ al. (2014). Best practices for scientific computing. PLoS
+ Biol. 12(1):e1001745.
+* Sandve GK, Nekrutenko A, Taylor J, Hovig E (2013) Ten Simple Rules
+ for Reproducible Computational Research. PLoS Comput Biol 9(10):
+ e1003285.
+* Alsheikh-Ali, Qureshi W, Al-Mallah MH, Ioannidis JP. (2013). Public
+ availability of published research data in high-impact
+ journals. PLoS
+ One. 6(9):e24357.
+* Laine C, Goodman SN, Griswold ME, Sox HC. (2007). Reproducible
+ research: moving toward research the public can really trust. Ann
+ Intern Med. 146(6):450-3. (Closed access)
+* Peng RD, Dominici F, Zeger SL (2006). Reproducible epidemiologic
+ research. Am J
+ Epidemiol. 163(9):783-789.
+* *(an informative blog)*
+*
+ *(Announcement of PLOS ONE 2012 Reproducibility Initiative)*
+
+# Git #
+
+* [Visual guide to Git](http://marklodato.github.io/visual-git-guide/index-en.html)
+* [Official Git documentation and introduction](http://git-scm.com/doc)
+* [Other documentation (nice and clean)](https://www.atlassian.com/git/tutorials)
+* [Hands-on tutorial, with web-based terminal](https://try.github.io/levels/1/challenges/1)
+* [A tutorial from a scientists perspective](http://nyuccl.org/pages/gittutorial/)
+* [An introduction to Git for researchers](http://datapub.cdlib.org/2014/05/05/github-a-primer-for-researchers/)
+* [Software carpentry lesson material for Git](http://swcarpentry.github.io/git-novice/)
+* [A scientists arguments and evidence for using Git](http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3639880/)
+* [Simpler first-steps guide](http://rogerdudler.github.io/git-guide/)
+* [Reference pages for all git commands](http://gitref.org/)
+* [Interactive, visual tutorial on branching](http://pcottle.github.io/learnGitBranching/)
+* [StackOverflow questions and answers on Git](http://stackoverflow.com/questions/tagged/git)
+
+# SAS ODS #
+
+* [SAS ODS Output Support](http://support.sas.com/documentation/cdl/en/odsug/65308/HTML/default/viewer.htm#p0oxrbinw6fjuwn1x23qam6dntyd.htm)
+*
+
+# Reproducible research #
+
+*
+*
+*
+*
+*
+*
+
+# Other resources #
+
+*
+*
+*
+
+# R #
+
+* [RStudio Cheat Sheets](https://www.rstudio.com/resources/cheatsheets/)
+
+## Data wrangling ##
+
+* [Common and frequently used commands in R](http://adv-r.had.co.nz/Vocabulary.html)
+* [Data frames in `dplyr`](https://cran.r-project.org/web/packages/dplyr/vignettes/data_frames.html)
+* [Introduction to `dplyr`](https://cran.r-project.org/web/packages/dplyr/vignettes/introduction.html)
+* [Cheatsheet for `dplyr` and `tidyr`](https://www.rstudio.com/wp-content/uploads/2015/02/data-wrangling-cheatsheet.pdf)
+* [Merging two dataframes using `dplyr`](https://cran.r-project.org/web/packages/dplyr/vignettes/two-table.html)
+* [Introduction to `tidyr`](https://cran.r-project.org/web/packages/tidyr/vignettes/tidy-data.html)
+* [Shorter intro to `tidyr`](http://blog.rstudio.org/2014/07/22/introducing-tidyr/)
+* [Regular expressions](http://www.regular-expressions.info/)
+* [Regular expression symbol meaning](http://www.endmemo.com/program/R/gsub.php)
+
+## R Markdown ##
+
+**Short guides/help:**
+
+* [R Markdown Cheat Sheet](http://www.rstudio.com/wp-content/uploads/2015/02/rmarkdown-cheatsheet.pdf)
+* [Markdown basics](http://rmarkdown.rstudio.com/authoring_basics.html)
+* [Code chunk options brief](http://rmarkdown.rstudio.com/authoring_rcodechunks.html)
+* [Bibliographies and citations](http://rmarkdown.rstudio.com/authoring_bibliographies_and_citations.html)
+* [Creating HTML documents (`YAML` options)](http://rmarkdown.rstudio.com/html_document_format.html)
+* [Creating Word documents (`YAML` options)](http://rmarkdown.rstudio.com/word_document_format.html)
+* [Using the `captioner` package](https://cran.r-project.org/web/packages/captioner/vignettes/using_captioner.html)
+
+**Detailed guides/help:**
+
+* [R Markdown site](http://rmarkdown.rstudio.com/)
+* [R Markdown more complete reference sheet](http://www.rstudio.com/wp-content/uploads/2015/03/rmarkdown-reference.pdf)
+* [More detailed markdown reference (`pandoc`)](http://rmarkdown.rstudio.com/authoring_pandoc_markdown.html)
+* [Pandoc Markdown website](http://pandoc.org/README.html)
+* [`knitr` website](http://yihui.name/knitr/)
+* [Code chunk options detailed](http://yihui.name/knitr/options/)
+* [Customization of markdown tables (`pander`)](http://rapporter.github.io/pander/)
+* [For installation or documentation of LaTeX](https://www.latex-project.org/)
diff --git a/lessons/rintro/assignment.md b/lessons/rintro/assignment.md
new file mode 100644
index 0000000..ae878bf
--- /dev/null
+++ b/lessons/rintro/assignment.md
@@ -0,0 +1,70 @@
+---
+title: "Assignment: Intro to R"
+author: Sarah Meister
+date: 2015-11-02
+layout: page
+sidebar: false
+tag:
+ - Lessons
+ - Assignment
+ - IntroR
+ - R
+categories:
+ - Lessons
+ - IntroR
+ - R
+output: html_vignette
+---
+
+# Intro to R Assignment #
+
+1. Create the following vectors in R:
+
+* a = 1 to 10
+* b = 11 to 20
+
+Use vector arithmetic to multiply these vectors and call the result c.
+
+Select subsets of c to identify the following:
+
+* What is the 5th element of c?
+* What are all of the elements of c which are greater than 50?
+
+2. Use R to create the following two matrices and multiply them. Make a new object of the resulting matrix (choose your own object name!)
+
+alpha:
+
+
+ {% highlight text %}
+ ## Error in kable_markdown(x = structure(c("7", "9", "12", "2", "4", "13": the table must have a header (column names)
+ {% endhighlight %}
+
+beta:
+
+
+ {% highlight text %}
+ ## Error in kable_markdown(x = structure(c("1", "7", "12", "19", "2", "8", : the table must have a header (column names)
+ {% endhighlight %}
+
+3. Load the `datasets` package and make a new dataframe with the `mtcars` dataset. What are the means of the columns? Hint: use a loop function.
+
+ Subset the `mtcars` dataset and make new vectors with:
+
+ * the column mpg
+ * the fourth column
+ * the third and fifth row
+ * hp is greater than 100
+
+4. Make a vector with the following arguments: "cat", "dog", "cow", "pig". Make a combination for loop and if/else expression that prints the second argument.
+
+5. Create a function called `stats` that takes two arguments and prints the mean and standard deviation of the first argument and the median of the second argument.
+
+ Hint: use the built-in `mean()`, `median()`, `sd()` and `print()` functions
+
+ Use your function with the `disp` and `drat` columns from the `mtcars` dataset.
+
+6. Use the `lapply()` function to find the range of all the columns in the `mtcars` dataframe. Now use `sapply()` and explain the difference between the outcomes as a comment in the script.
+
+7. Use `mapply()` to find the sum of the `mpg`, `wt` and `qsec` columns of the mtcars dataframe.
+
+8. Use the `tapply()` function to apply the `max()` function on the `hp` column by the `gear` column.
diff --git a/lessons/rintro/assignmentAnswers.md b/lessons/rintro/assignmentAnswers.md
new file mode 100644
index 0000000..0cef3b0
--- /dev/null
+++ b/lessons/rintro/assignmentAnswers.md
@@ -0,0 +1,371 @@
+---
+published: true
+---
+
+1.
+
+
+{% highlight r %}
+a <- c(1:10)
+b <- c(11:20)
+
+(c <- a*b)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 11 24 39 56 75 96 119 144 171 200
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+c[5]
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 75
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+subset(c, c > 50)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 56 75 96 119 144 171 200
+{% endhighlight %}
+
+
+2.
+
+{% highlight r %}
+(d <- matrix(c(7,9,12,2,4,13), 2, 3))
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [,1] [,2] [,3]
+## [1,] 7 12 4
+## [2,] 9 2 13
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+(e <- matrix(c(1,7,12,19,2,8,13,20,3,9,14,21), 3, 4))
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [,1] [,2] [,3] [,4]
+## [1,] 1 19 13 9
+## [2,] 7 2 20 14
+## [3,] 12 8 3 21
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+
+(f <- d %*% e)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [,1] [,2] [,3] [,4]
+## [1,] 139 189 343 315
+## [2,] 179 279 196 382
+{% endhighlight %}
+
+3.
+
+
+{% highlight r %}
+library(datasets)
+
+(g <- mtcars)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## mpg cyl disp hp drat wt qsec vs am gear carb
+## Mazda RX4 21.0 6 160.0 110 3.90 2.620 16.46 0 1 4 4
+## Mazda RX4 Wag 21.0 6 160.0 110 3.90 2.875 17.02 0 1 4 4
+## Datsun 710 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1
+## Hornet 4 Drive 21.4 6 258.0 110 3.08 3.215 19.44 1 0 3 1
+## Hornet Sportabout 18.7 8 360.0 175 3.15 3.440 17.02 0 0 3 2
+## Valiant 18.1 6 225.0 105 2.76 3.460 20.22 1 0 3 1
+## Duster 360 14.3 8 360.0 245 3.21 3.570 15.84 0 0 3 4
+## Merc 240D 24.4 4 146.7 62 3.69 3.190 20.00 1 0 4 2
+## Merc 230 22.8 4 140.8 95 3.92 3.150 22.90 1 0 4 2
+## Merc 280 19.2 6 167.6 123 3.92 3.440 18.30 1 0 4 4
+## Merc 280C 17.8 6 167.6 123 3.92 3.440 18.90 1 0 4 4
+## Merc 450SE 16.4 8 275.8 180 3.07 4.070 17.40 0 0 3 3
+## Merc 450SL 17.3 8 275.8 180 3.07 3.730 17.60 0 0 3 3
+## Merc 450SLC 15.2 8 275.8 180 3.07 3.780 18.00 0 0 3 3
+## Cadillac Fleetwood 10.4 8 472.0 205 2.93 5.250 17.98 0 0 3 4
+## Lincoln Continental 10.4 8 460.0 215 3.00 5.424 17.82 0 0 3 4
+## Chrysler Imperial 14.7 8 440.0 230 3.23 5.345 17.42 0 0 3 4
+## Fiat 128 32.4 4 78.7 66 4.08 2.200 19.47 1 1 4 1
+## Honda Civic 30.4 4 75.7 52 4.93 1.615 18.52 1 1 4 2
+## Toyota Corolla 33.9 4 71.1 65 4.22 1.835 19.90 1 1 4 1
+## Toyota Corona 21.5 4 120.1 97 3.70 2.465 20.01 1 0 3 1
+## Dodge Challenger 15.5 8 318.0 150 2.76 3.520 16.87 0 0 3 2
+## AMC Javelin 15.2 8 304.0 150 3.15 3.435 17.30 0 0 3 2
+## Camaro Z28 13.3 8 350.0 245 3.73 3.840 15.41 0 0 3 4
+## Pontiac Firebird 19.2 8 400.0 175 3.08 3.845 17.05 0 0 3 2
+## Fiat X1-9 27.3 4 79.0 66 4.08 1.935 18.90 1 1 4 1
+## Porsche 914-2 26.0 4 120.3 91 4.43 2.140 16.70 0 1 5 2
+## Lotus Europa 30.4 4 95.1 113 3.77 1.513 16.90 1 1 5 2
+## Ford Pantera L 15.8 8 351.0 264 4.22 3.170 14.50 0 1 5 4
+## Ferrari Dino 19.7 6 145.0 175 3.62 2.770 15.50 0 1 5 6
+## Maserati Bora 15.0 8 301.0 335 3.54 3.570 14.60 0 1 5 8
+## Volvo 142E 21.4 4 121.0 109 4.11 2.780 18.60 1 1 4 2
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+apply(g, 2, mean)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## mpg cyl disp hp drat wt
+## 20.090625 6.187500 230.721875 146.687500 3.596563 3.217250
+## qsec vs am gear carb
+## 17.848750 0.437500 0.406250 3.687500 2.812500
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+(h <- g[,"mpg"])
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 21.0 21.0 22.8 21.4 18.7 18.1 14.3 24.4 22.8 19.2 17.8 16.4 17.3 15.2
+## [15] 10.4 10.4 14.7 32.4 30.4 33.9 21.5 15.5 15.2 13.3 19.2 27.3 26.0 30.4
+## [29] 15.8 19.7 15.0 21.4
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+#OR
+
+(h <- g$mpg)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 21.0 21.0 22.8 21.4 18.7 18.1 14.3 24.4 22.8 19.2 17.8 16.4 17.3 15.2
+## [15] 10.4 10.4 14.7 32.4 30.4 33.9 21.5 15.5 15.2 13.3 19.2 27.3 26.0 30.4
+## [29] 15.8 19.7 15.0 21.4
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+(i <- g[, 4])
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 110 110 93 110 175 105 245 62 95 123 123 180 180 180 205 215 230
+## [18] 66 52 65 97 150 150 245 175 66 91 113 264 175 335 109
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+(j <- g[c(3,5),])
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## mpg cyl disp hp drat wt qsec vs am gear carb
+## Datsun 710 22.8 4 108 93 3.85 2.32 18.61 1 1 4 1
+## Hornet Sportabout 18.7 8 360 175 3.15 3.44 17.02 0 0 3 2
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+(k <- subset(g, g$hp < 100))
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## mpg cyl disp hp drat wt qsec vs am gear carb
+## Datsun 710 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1
+## Merc 240D 24.4 4 146.7 62 3.69 3.190 20.00 1 0 4 2
+## Merc 230 22.8 4 140.8 95 3.92 3.150 22.90 1 0 4 2
+## Fiat 128 32.4 4 78.7 66 4.08 2.200 19.47 1 1 4 1
+## Honda Civic 30.4 4 75.7 52 4.93 1.615 18.52 1 1 4 2
+## Toyota Corolla 33.9 4 71.1 65 4.22 1.835 19.90 1 1 4 1
+## Toyota Corona 21.5 4 120.1 97 3.70 2.465 20.01 1 0 3 1
+## Fiat X1-9 27.3 4 79.0 66 4.08 1.935 18.90 1 1 4 1
+## Porsche 914-2 26.0 4 120.3 91 4.43 2.140 16.70 0 1 5 2
+{% endhighlight %}
+
+4.
+
+
+{% highlight r %}
+
+ani <- c("cat", "dog", "cow", "pig")
+for (i in ani) {
+ if (i == "dog") {
+ print(i) }
+ else {
+ }
+}
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] "dog"
+{% endhighlight %}
+
+5.
+
+
+{% highlight r %}
+stats <- function(a,b) {
+ print (mean(a))
+ print (sd(a))
+ print(median(b))
+}
+
+stats(g$disp, g$drat)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 230.7219
+## [1] 123.9387
+## [1] 3.695
+{% endhighlight %}
+
+6.
+
+{% highlight r %}
+lapply(g, range, na.rm = TRUE)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## $mpg
+## [1] 10.4 33.9
+##
+## $cyl
+## [1] 4 8
+##
+## $disp
+## [1] 71.1 472.0
+##
+## $hp
+## [1] 52 335
+##
+## $drat
+## [1] 2.76 4.93
+##
+## $wt
+## [1] 1.513 5.424
+##
+## $qsec
+## [1] 14.5 22.9
+##
+## $vs
+## [1] 0 1
+##
+## $am
+## [1] 0 1
+##
+## $gear
+## [1] 3 5
+##
+## $carb
+## [1] 1 8
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+sapply(g, range, na.rm = TRUE)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## mpg cyl disp hp drat wt qsec vs am gear carb
+## [1,] 10.4 4 71.1 52 2.76 1.513 14.5 0 0 3 1
+## [2,] 33.9 8 472.0 335 4.93 5.424 22.9 1 1 5 8
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+## lapply always returns a list while sapply simiplied the result and returned a matrix
+{% endhighlight %}
+
+7.
+
+{% highlight r %}
+mapply(range, mtcars[,c(1,6,7)], na.rm = TRUE)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## mpg wt qsec
+## [1,] 10.4 1.513 14.5
+## [2,] 33.9 5.424 22.9
+{% endhighlight %}
+
+8.
+
+{% highlight r %}
+tapply(mtcars$hp, mtcars$gear, max, na.rm = TRUE)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## 3 4 5
+## 245 123 335
+{% endhighlight %}
diff --git a/lessons/rintro/cheatsheet.md b/lessons/rintro/cheatsheet.md
new file mode 100644
index 0000000..a187523
--- /dev/null
+++ b/lessons/rintro/cheatsheet.md
@@ -0,0 +1,1079 @@
+---
+title: "Cheatsheet: Introduction to R"
+author:
+ - Sarah Meister
+date: 2015-11-02
+layout: page
+sidebar: false
+tag:
+ - Lessons
+ - Cheatsheet
+ - IntroR
+ - R
+categories:
+ - Lessons
+ - IntroR
+ - R
+---
+
+Brief description/intro
+
+# (language) commands: Some useful or common ones #
+
+## `command` ##
+
+> Description
+
+> Example code:
+
+ example code (note the tab/4 spaces indent)
+
+## `command` ##
+
+> Description
+
+> Example code:
+
+ example code (note the tab/4 spaces indent)
+
+R is an object-oriented programing language, meaning everything we encounter in R is an object.
+
+R has several types of objects:
+
+R has a 5 basic classes of objects:
+
+* Character
+* Numeric
+* Integer
+* Complex
+* Logical
+
+R has 5 different types of objects:
+
+* Vectors
+* Lists
+* Matrices
+* Factors
+* Dataframes
+
+Thus you could have a logical vector, a character dataframe, a numeric matrix, etc.
+
+# Vectors #
+
+The most basic object is a vector. A vector is a sequence of objects of the same class. We can make vectors using the `c()` function.
+
+
+{% highlight r %}
+a <- c(0.4, 1.2) # numeric
+b <- c(TRUE, FALSE) # logical
+c <- c("a","b","c") # character
+d <- c(1L, 2L) # integer
+e <- 1:10 # integer
+f <- c(1+0i, 2+4i) # complex
+{% endhighlight %}
+
+Empty vectors can be made using the `vector()` function
+
+
+{% highlight r %}
+(x <- vector("numeric", length=10))
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 0 0 0 0 0 0 0 0 0 0
+{% endhighlight %}
+
+# Lists #
+
+A list is a special type of vector that can contain objects of different classes. For instance, we can have a list with a numeric dataframe as one object within the list, and a string vector as another.
+
+
+{% highlight r %}
+(x <- list(1,"a", TRUE, 1+4i))
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [[1]]
+## [1] 1
+##
+## [[2]]
+## [1] "a"
+##
+## [[3]]
+## [1] TRUE
+##
+## [[4]]
+## [1] 1+4i
+{% endhighlight %}
+
+The elements of a list are seperated by double brackets.
+
+# Matrices #
+
+Matrices are vectors with a dimension attribute. The dim attribute is an integer vector of length = 2 (nrow, ncol). Matrix multiplication is done with the %*% operator
+
+
+{% highlight r %}
+(x <- matrix(1:6, nrow=2, ncol=3))
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [,1] [,2] [,3]
+## [1,] 1 3 5
+## [2,] 2 4 6
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+dim(x)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 2 3
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+attributes(x)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## $dim
+## [1] 2 3
+{% endhighlight %}
+
+Matrices are constructed column-wise, so entries start at the upper left corner and run down the columns.
+
+Matrices can be created directly from vectors by adding a dimension value
+
+
+{% highlight r %}
+(x <- 1:10)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 1 2 3 4 5 6 7 8 9 10
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+(dim(x) <- c(2,5))
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 2 5
+{% endhighlight %}
+
+We can also create matrices by column-binding or row-binding with the functions `cbind()` and `rbind()`
+
+
+{% highlight r %}
+x <- 1:3
+y <- 10:12
+cbind(x,y)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## x y
+## [1,] 1 10
+## [2,] 2 11
+## [3,] 3 12
+{% endhighlight %}
+
+
+
+{% highlight r %}
+rbind (x, y)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [,1] [,2] [,3]
+## x 1 2 3
+## y 10 11 12
+{% endhighlight %}
+
+# Factors #
+
+Factors are used to represent catagorical data. They can be ordered or unordered.
+
+* They are like an integer vector where each integer has a label.
+* Factors are more descriptive than integers e.g. "Male" and "Female" vs. 1 and 2
+* Factors are treated specially by modelling functions like `lm()` and `glm()`
+
+
+{% highlight r %}
+(x <- factor(c("yes", "yes", "no", "no", "yes")))
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] yes yes no no yes
+## Levels: no yes
+{% endhighlight %}
+
+
+
+{% highlight r %}
+table (x)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## x
+## no yes
+## 2 3
+{% endhighlight %}
+
+
+
+{% highlight r %}
+unclass(x)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 2 2 1 1 2
+## attr(,"levels")
+## [1] "no" "yes"
+{% endhighlight %}
+
+# Dataframe #
+
+Dataframes are used in R to store tabular data. They are represented in R as a special type of list where every element in the list has the same length. Each element of the list can be thought of as a **column** and each length of the list can be thought of as a **row**. Because a dataframe is a type of list, dataframes can store different classes of objects in each column (e.g. numeric, character, logical, etc.).
+
+Depending on the source of the data, dataframes can be made using the `read.table()`, `read.csv` or `data.frame()` functions.
+
+
+{% highlight r %}
+(x <- data.frame(foo = 1:4, bar = c(T,T,F,F)))
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## foo bar
+## 1 1 TRUE
+## 2 2 TRUE
+## 3 3 FALSE
+## 4 4 FALSE
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+nrow(x)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 4
+{% endhighlight %}
+
+
+
+{% highlight r %}
+ncol(x)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 2
+{% endhighlight %}
+
+
+# Attributes #
+
+R objects can have attributes, such as:
+
+* names
+* classes
+* lengths
+* dimensions (matrices/dataframes)
+* user defined attributes, such as metadata
+* dataframes have a special attribute called `row.names`
+
+Attributes can be modified with the `attributes()` function, among others.
+
+For example, modifiying the `names` of a vector can make your code more readable.
+
+{% highlight r %}
+x <- 1:3
+names(x)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## NULL
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+names(x) <- c("alpha", "beta", "gamma")
+
+x
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## alpha beta gamma
+## 1 2 3
+{% endhighlight %}
+
+
+
+{% highlight r %}
+names(x)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] "alpha" "beta" "gamma"
+{% endhighlight %}
+
+# Looking for help in R #
+
+* Look up the documentation for a function: `help("function")` OR
+
+* ?function
+
+* Look up documentation for a package: `help(package="packagename")`
+
+# Packages #
+
+R comes with a few default packages, however there are thousands of R packages that extend R's capabilities (such as `ggplot2`, our next lesson)
+
+* To see what packages are loaded: `search()`
+
+* To view available packages: `library()`
+
+* To load a package: `library(packagename)`
+
+* Install new package: `install.packages("packagename")`
+
+# Working Directory #
+
+R knows the directory it was started in, and refers to this as the "working directory". This is where R will search for any files you are imputting, and where it will place any files you output.
+
+To look at the current working directory: `getwd()`
+
+To change the working directory: ` setwd("directory_path")`
+
+To make a new folder in R: `dir.create(path)`
+
+# Imputting Tabular Data #
+
+Most data you will work with comes in tabular form (such as an Excel spreadsheet). The two most commonly-used functions used to imput tabular data into R are `read.table()` and `read.csv()`.
+
+{% highlight r %}
+data <- read.table("file_name.txt", header=FALSE, sep="\t")
+
+# \t is the regular expression for the tab key. This means our table is tab-delimited (tab-separated)
+
+data2 <- read.csv("file_name.csv", header=TRUE)
+
+#.csv is a comma-separated table
+{% endhighlight %}
+
+# Outputting Tabular Data #
+
+Often you'll want to save R data into a tabular form (e.g. to view in Excel). This is where you use `write.table()`
+
+{% highlight r %}
+write.table(data, file= "myfile.csv", sep=",", row.names=TRUE, col.names=FALSE)
+{% endhighlight %}
+
+# Subsetting Data #
+
+Using `[]` in R allows you to subset certain parts of a vector
+
+
+{% highlight r %}
+x <- c(TRUE, FALSE, "alpha", "beta", "gamma", 1, 2)
+
+x[c(1, 4, 5)]
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] "TRUE" "beta" "gamma"
+{% endhighlight %}
+
+This also works for matrices
+
+
+{% highlight r %}
+a <- matrix(1:9, nrow = 3)
+colnames(a) <- c("A", "B", "C")
+a[1:2, ]
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## A B C
+## [1,] 1 4 7
+## [2,] 2 5 8
+{% endhighlight %}
+
+
+
+{% highlight r %}
+a[c(T, F, T), c("B", "A")]
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## B A
+## [1,] 4 1
+## [2,] 6 3
+{% endhighlight %}
+
+
+
+{% highlight r %}
+a[0, -2]
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## A C
+{% endhighlight %}
+
+and dataframes
+
+
+{% highlight r %}
+df <- data.frame(x = 1:3, y = 3:1, z = letters[1:3])
+
+df[ ,c(1, 3)] #gives the columns that are in position 1 and 3
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## x z
+## 1 1 a
+## 2 2 b
+## 3 3 c
+{% endhighlight %}
+
+You can also easily subset a column in a dataframe using a $
+
+
+{% highlight r %}
+df <- data.frame(x = 1:10, y = 11:20, z = letters[1:10])
+
+df$z
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] a b c d e f g h i j
+## Levels: a b c d e f g h i j
+{% endhighlight %}
+
+# R Operators #
+
+Assignment operators
+
+|Operator | Description|
+|---- | ---- |
+| <- | local environment assignment |
+| <<- | global environment assignment |
+
+Arthmetic Operators
+
+|Operator | Description|
+|-------- | -----------|
+|+ | addition|
+|- | subtraction|
+|* | multiplication|
+|/ | division|
+|^ or ** | exponentiation|
+|x %% y | modulus (5 %% 2 is 1)|
+|x %/% y | integer division (5%/% is 2)|
+|x %*% y | matrix multiplication|
+
+Logical Operators
+
+Operator | Description
+-------- | -----------
+< | less than
+<= | less than or equal to
+> | greater than
+>= | greater than or equal to
+== | exactly equal to
+!= | not equal to
+!x | not x
+x | y | x OR y (returns a vector)
+x || y | x OR y (examines only 1st element of vector)
+x & y | x AND y (returns a vector)
+x && y | x AND y (examines only 1st element of vector)
+isTRUE(x) | test if X is TRUE
+
+## Commenting in R #
+
+
+{% highlight r %}
+# commenting in R is easy! Everything past a # means that R will ignore whatever is written there.
+{% endhighlight %}
+
+# subset function #
+
+The `subset()` function in R will return a subset of an argument that meet a particular condition.
+
+
+{% highlight r %}
+numvec = c(2,5,8,9,0,6,7,8,4,5,7,11)
+
+subset(numvec, numvec < 9 & numvec > 4)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 5 8 6 7 8 5 7
+{% endhighlight %}
+
+# if-else expressions #
+
+
+{% highlight r %}
+if (condition) {
+ ##do this
+} else {
+ ##do that
+}
+
+## to add more conditions
+
+if (condition1) {
+ ## do this
+} else if (condition2) {
+ ## do something different
+} else {
+ ## do something else
+}
+{% endhighlight %}
+
+# if-else expressions #
+
+
+{% highlight r %}
+if (x > 3) {
+ y <- 10
+} else {
+ y <- 0
+}
+
+
+### OR
+
+
+y <- if(x > 3) {
+ 10
+} else {
+ 0
+}
+{% endhighlight %}
+
+# for loop #
+
+{% highlight r %}
+for (i in 1:10) {
+ print (i)
+}
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 1
+## [1] 2
+## [1] 3
+## [1] 4
+## [1] 5
+## [1] 6
+## [1] 7
+## [1] 8
+## [1] 9
+## [1] 10
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+
+### nested for loops
+
+x <- matrix(1:6, 2, 3)
+
+for (i in seq_len(nrow(x))) {
+ for (j in seq_len(ncol(x))) {
+ print (x[i, j])
+ }
+}
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 1
+## [1] 3
+## [1] 5
+## [1] 2
+## [1] 4
+## [1] 6
+{% endhighlight %}
+
+# Writing functions #
+
+functions are created using the `function()` directive and are stored as R objects of the class "function". Functions can be passed as arguments to other functions and they can be nested (functions inside functions)
+
+
+{% highlight r %}
+f <- function(argument) {
+ ## do something here
+}
+{% endhighlight %}
+
+
+{% highlight r %}
+
+## make a function called above10 with the argument x that subsets all variables of x that are greater than 10
+above10 <- function(x) {
+ use <- x > 10
+ x[use]
+}
+
+## make a function called above with the arguments x & n that subsets all variables of x that are greater than n
+above <- function(x, n) {
+ use <- x > n
+ x[use]
+}
+
+## create a function called columnmean that takes the arguments y and the argument remove.NA with a default value of TRUE. This function takes the number of columns of y, turns it into a numeric object, and calculates the mean of each column of y while removing NA values
+columnmean <- function (y, remove.NA = TRUE) {
+ nc <- ncol(y)
+ means <- numeric(nc)
+ for (i in 1:nc) {
+ means[i] <- mean(y[,i], na.rm = remove.NA)
+ }
+ means
+}
+{% endhighlight %}
+# Loop Functions #
+
+R has built loop functions, which often have the word "apply" in them.
+
+- `lapply`: loop over a list and evaluate a function on each element
+- `sapply`: same as `lapply` but it tries to simplify the result
+- `apply`: apply a function over the margins of an array
+- `tapply`: apply a function over subsets of a vector (`lapply()` used with `split()` does the same thing)
+- `mapply`: a multivariate version of `lapply`
+
+# lapply #
+
+`lapply` loops internally with C code, making the looping faster
+
+`lapply` takes two to three arguments:
+
+1. a list `x` (or another object that is coerced to a list)
+2. a function
+3. other arguments as necessary
+
+
+{% highlight r %}
+x <- list(a=1:5, b=rnorm(10), c=rnorm(20,1), d=rnorm(100,5))
+lapply(x, mean)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## $a
+## [1] 3
+##
+## $b
+## [1] 0.01662391
+##
+## $c
+## [1] 0.9641099
+##
+## $d
+## [1] 5.239093
+{% endhighlight %}
+`lapply` will always return a list
+
+`lapply` and others make heavy use of anonymous functions, which are functions without names.
+
+{% highlight r %}
+x <- list(a=matrix(1:4,2,2), b=matrix(1:6,3,2))
+x
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## $a
+## [,1] [,2]
+## [1,] 1 3
+## [2,] 2 4
+##
+## $b
+## [,1] [,2]
+## [1,] 1 4
+## [2,] 2 5
+## [3,] 3 6
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+lapply(x, function(ele) ele[ ,1])
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## $a
+## [1] 1 2
+##
+## $b
+## [1] 1 2 3
+{% endhighlight %}
+
+
+
+{% highlight r %}
+## create an anonymous function with the argument ele and then define that argument.
+{% endhighlight %}
+
+# sapply #
+
+`sapply` will try to simplify the result of lapply if possible
+
+- if the result is a list where every element is length 1, than a vector is returned
+- if the result is a list where every element is a vector of the same length, a matrix is returned
+- otherwise a list is returned
+
+
+{% highlight r %}
+x <- list(a=1:5, b=rnorm(10), c=rnorm(20,1), d=rnorm(100,5))
+sapply(x, mean)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## a b c d
+## 3.0000000 0.1447111 1.4409702 4.9880345
+{% endhighlight %}
+
+# apply #
+
+`apply` is used to evalute a function over the margins of an array. It is not faster than a loop, but it works in one line. `1` means do the function to the rows. `2` means do the function to the columns
+
+
+{% highlight r %}
+x <- matrix (rnorm(200), 20, 10)
+apply(x, 2, mean) # keeping number of columns and collapsing rows. This gives a vector of the means of columns.
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] -0.11571236 -0.06699980 0.01217097 0.44350588 0.19404572
+## [6] 0.40123394 0.06988406 -0.24671559 -0.18478261 -0.34146637
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+apply(x, 1, sum) # this calculates the sum of all rows
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 3.0559833 2.2761056 -0.7529187 -1.7810694 0.3799718 4.1971014
+## [7] -3.7791267 -1.1373524 -0.5365995 2.5774285 -1.4991704 0.4736595
+## [13] 0.4518346 -3.8727141 6.6928887 1.9541773 -4.5425821 -0.6554135
+## [19] 0.5273807 -0.7263079
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+y <- matrix(rnorm(200), 20, 10)
+
+apply(y, 1, quantile, probs = c(0.25, 0.75))
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [,1] [,2] [,3] [,4] [,5] [,6]
+## 25% -0.5016427 -1.1950471 -0.2632323 -0.72541103 -0.8288047 -0.1112117
+## 75% 0.6660713 -0.1398314 0.1706738 -0.03855607 0.7696287 0.4328380
+## [,7] [,8] [,9] [,10] [,11] [,12]
+## 25% -0.3790344 -0.0007273004 -0.5932285 -1.0311000 -0.5114152 -1.05972346
+## 75% 0.4960863 0.9409016018 0.6469670 0.8245869 0.4520464 -0.09879784
+## [,13] [,14] [,15] [,16] [,17] [,18]
+## 25% -0.2181878 -0.09879262 -0.6041801 -0.3944707 -0.9776292 0.09703779
+## 75% 0.9720584 0.34848220 0.7378028 0.2837990 0.4302688 1.02127401
+## [,19] [,20]
+## 25% -0.4975348 -0.8437645
+## 75% 0.1902392 0.6093546
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+# with an array (stacks of matrices: multidimensional)
+
+a <- array(rnorm(2*2*10), c(2,2,10)) #create an array that looks like a bunch of 2 by 2 matrices and take the mean of those
+
+apply(a, c(1,2), mean) #this keeps the 1st and 2nd dimension
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [,1] [,2]
+## [1,] -0.32013553 -0.02170755
+## [2,] 0.06619694 0.24833397
+{% endhighlight %}
+
+# mapply #
+
+`mapply` is a multivariate apply (like `lapply`) that applies a function over a set of arguments.
+
+
+{% highlight r %}
+mapply(rep, 1:4, 4:1)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [[1]]
+## [1] 1 1 1 1
+##
+## [[2]]
+## [1] 2 2 2
+##
+## [[3]]
+## [1] 3 3
+##
+## [[4]]
+## [1] 4
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+# is the same as
+
+list(rep(1, 4), rep(2,3), rep(3,2), rep(4,1))
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [[1]]
+## [1] 1 1 1 1
+##
+## [[2]]
+## [1] 2 2 2
+##
+## [[3]]
+## [1] 3 3
+##
+## [[4]]
+## [1] 4
+{% endhighlight %}
+
+# Instant Vectorization to make your code faster#
+
+
+{% highlight r %}
+
+## create a function called noise that takes the arguments n, mean and sd and produces random varibles with those specifications.
+
+noise<- function(n,mean, sd) {
+ rnorm(n, mean, sd)
+}
+
+## apply the noise function with n=5, mean=1 and sd=2
+
+noise(5, 1, 2)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] -1.2807770 -0.3236428 1.1059255 -0.6843800 2.0478857
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+## apply the noise function for n= 1 to 5, mean = 1 to 5, and sd = 2
+
+mapply(noise, 1:5, 1:5, 2)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [[1]]
+## [1] -1.165767
+##
+## [[2]]
+## [1] 1.768898 2.595327
+##
+## [[3]]
+## [1] 5.439703 5.308744 5.027059
+##
+## [[4]]
+## [1] 6.4861905 2.5891246 0.8992601 3.2437817
+##
+## [[5]]
+## [1] 6.027180 4.904761 2.706628 3.730056 1.632191
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+## which is the same as writing
+
+list(noise(1,1,2), noise(2,2,2), noise(3,3,2), noise(4,4,2), noise(5,5,2))
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [[1]]
+## [1] 2.903614
+##
+## [[2]]
+## [1] 2.644925 -2.099117
+##
+## [[3]]
+## [1] 0.4289258 5.3264939 1.5264498
+##
+## [[4]]
+## [1] 6.643890 4.803451 3.420890 4.351345
+##
+## [[5]]
+## [1] 4.284911 4.091920 3.406254 7.343844 6.684944
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+# (the outputs are different because our function makes new random varibles each time)
+{% endhighlight %}
+
+# tapply #
+
+`tapply` is used to apply a function over subsets of a vector. It will simplify the result, like sapply.
+
+
+{% highlight r %}
+x <- c(1:30) # make a vector with 30 variables
+f<- gl(3,10) # make a factor variable with 3 levels and 10 reps of each level
+f
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 1 1 1 1 1 1 1 1 1 1 2 2 2 2 2 2 2 2 2 2 3 3 3 3 3 3 3 3 3 3
+## Levels: 1 2 3
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+tapply(x, f, mean) # apply the mean function to x, subsetting by f
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## 1 2 3
+## 5.5 15.5 25.5
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+tapply(x, f, range) # apply the range function to x, subsetting by f
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## $`1`
+## [1] 1 10
+##
+## $`2`
+## [1] 11 20
+##
+## $`3`
+## [1] 21 30
+{% endhighlight %}
+
+
diff --git a/lessons/rintro/intro.md b/lessons/rintro/intro.md
new file mode 100644
index 0000000..8cae602
--- /dev/null
+++ b/lessons/rintro/intro.md
@@ -0,0 +1,55 @@
+---
+title: "Introduction: R"
+author:
+ - Sarah Meister
+date: 2015-11-02
+layout: page
+sidebar: false
+tag:
+ - Lessons
+ - Introduction
+ - IntroR
+ - R
+categories:
+ - Lessons
+ - IntroR
+ - R
+permalink: lessons/git/
+---
+
+# What is R and why do I need to learn it? #
+
+R is an object-oritented programming language for statistical computing and graphics.
+
+Benefits of R:
+1. Easily extenible through **FREE** R packages
+2. Active community (lots on online help)
+3. Able to produce publication-quality graphs, and dynamic and interactive graphs with additional packages (did I mention they were **FREE**??)
+4. For computationally intensive tasks, R objects can be manipulated with C, C++, Java, .NET or Python code
+5. Has it's own flavor of Markdown, making R code easily documented and shareable (which you will be learning in another lesson)
+
+## Learning objectives ##
+
+After this workshop, our expectation is that you will be able to:
+
+1. Understand the "grammar" of the R language
+2. Import and export data into R
+3. View data in R and perform basic data calculations & manipulations
+4. Make and use a basic R function
+5. Understand and use the useful built-in loop functions
+
+# Materials for this lesson: #
+
+* [Slides](slides/)
+* [Cheatsheet](cheatsheet/)
+* [Assignment](assignment/)
+* [Assignment Answers](answer)
+
+Other resources can be found here:
+
+* [CRAN website](https://cran.r-project.org/)
+* [CRAN R Reference Card](https://cran.r-project.org/doc/contrib/Short-refcard.pdf)
+* [Interactive R tutorial](http://tryr.codeschool.com/)
+* [R in a Nutshell](http://web.udl.es/Biomath/Bioestadistica/R/Manuals/r_in_a_nutshell.pdf)
+
+
diff --git a/lessons/rintro/livecoding.md b/lessons/rintro/livecoding.md
new file mode 100644
index 0000000..4bdd1f4
--- /dev/null
+++ b/lessons/rintro/livecoding.md
@@ -0,0 +1,481 @@
+---
+title: "Introduction to R"
+author: "Sarah Meister"
+date: '2015-11-02'
+output: html_vignette
+layout: page
+sidebar: no
+published: true
+---
+
+# Starting R #
+
+We will be using R studio for this course as it's a user-friendly GUI for R. You can also access R from Terminal (Mac) or Command Line (Windows) if you please.
+
+
+{% highlight r %}
+getwd()
+
+setwd("/Users/thesarahmeister/")
+
+dir.create("Desktop/practice-2015-10/sarah/intror")
+
+setwd("Desktop/practice-2015-10/sarah/intror")
+
+dir()
+
+{% endhighlight %}
+
+# Now let's code! #
+
+Download the `airQuality.csv` file from our [Github repo](https://github.com/codeasmanuscript/practice-2015-10) and import it into R using the `read.csv` function
+
+
+
+{% highlight r %}
+airQuality <- read.csv("airQuality.csv", header = TRUE)
+{% endhighlight %}
+
+Now let's look at the data using a few different functions
+
+
+{% highlight r %}
+head(airQuality)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## Ozone Solar.R Wind Temp Month Day
+## 1 41 190 7.4 67 May 1
+## 2 36 118 8.0 72 May 2
+## 3 12 149 12.6 74 May 3
+## 4 18 313 11.5 62 May 4
+## 5 NA NA 14.3 56 May 5
+## 6 28 NA 14.9 66 May 6
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+head(airQuality, n=8)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## Ozone Solar.R Wind Temp Month Day
+## 1 41 190 7.4 67 May 1
+## 2 36 118 8.0 72 May 2
+## 3 12 149 12.6 74 May 3
+## 4 18 313 11.5 62 May 4
+## 5 NA NA 14.3 56 May 5
+## 6 28 NA 14.9 66 May 6
+## 7 23 299 8.6 65 May 7
+## 8 19 99 13.8 59 May 8
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+tail(airQuality)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## Ozone Solar.R Wind Temp Month Day
+## 148 14 20 16.6 63 Sep 25
+## 149 30 193 6.9 70 Sep 26
+## 150 NA 145 13.2 77 Sep 27
+## 151 14 191 14.3 75 Sep 28
+## 152 18 131 8.0 76 Sep 29
+## 153 20 223 11.5 68 Sep 30
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+str(airQuality)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## 'data.frame': 153 obs. of 6 variables:
+## $ Ozone : int 41 36 12 18 NA 28 23 19 8 NA ...
+## $ Solar.R: int 190 118 149 313 NA NA 299 99 19 194 ...
+## $ Wind : num 7.4 8 12.6 11.5 14.3 14.9 8.6 13.8 20.1 8.6 ...
+## $ Temp : int 67 72 74 62 56 66 65 59 61 69 ...
+## $ Month : Factor w/ 5 levels "Aug","Jul","Jun",..: 4 4 4 4 4 4 4 4 4 4 ...
+## $ Day : int 1 2 3 4 5 6 7 8 9 10 ...
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+summary(airQuality)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## Ozone Solar.R Wind Temp
+## Min. : 1.00 Min. : 7.0 Min. : 1.700 Min. :56.00
+## 1st Qu.: 18.00 1st Qu.:115.8 1st Qu.: 7.400 1st Qu.:72.00
+## Median : 31.50 Median :205.0 Median : 9.700 Median :79.00
+## Mean : 42.13 Mean :185.9 Mean : 9.958 Mean :77.88
+## 3rd Qu.: 63.25 3rd Qu.:258.8 3rd Qu.:11.500 3rd Qu.:85.00
+## Max. :168.00 Max. :334.0 Max. :20.700 Max. :97.00
+## NA's :37 NA's :7
+## Month Day
+## Aug:31 Min. : 1.0
+## Jul:31 1st Qu.: 8.0
+## Jun:30 Median :16.0
+## May:31 Mean :15.8
+## Sep:30 3rd Qu.:23.0
+## Max. :31.0
+##
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+nrow(airQuality)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 153
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+ncol(airQuality)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 6
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+range(airQuality$Temp)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 56 97
+{% endhighlight %}
+
+Lets alter our dataset a bit
+
+
+{% highlight r %}
+
+names(airQuality$Solar.R) <- "Solar"
+
+airQuality$Random <- rnorm(153, 1, 2)
+{% endhighlight %}
+
+Okay time to subset our data
+
+
+{% highlight r %}
+
+rowThree <- airQuality[3, ]
+
+colFour <- airQuality[ ,4]
+
+ozone <- airQuality$Ozone
+
+highTemp <- airQuality[airQuality$Temp > 80, ]
+
+may <- airQuality[airQuality$Month == "May", ]
+
+highTempOrMay <- airQuality[airQuality$Month == "May" | airQuality$Temp > 80, ]
+
+lowTempAndSep <- subset(airQuality, airQuality$Month == "Sep" & airQuality$Temp < 70)
+{% endhighlight %}
+
+okay let's perform some basic data manipulations on our dataframe and subsetted data
+
+
+{% highlight r %}
+
+mean(airQuality$Ozone) #returns an NA
+
+mean(airQuality$Ozone, na.rm=TRUE)
+
+colMeans(airQuality, na.rm=TRUE) #this one doesn't work
+
+airQualNoMonth <- airQuality[,-5]
+
+colMeans(airQualNoMonth, na.rm=TRUE)
+
+sd(colFour)
+
+sd(airQualNoMonth)
+{% endhighlight %}
+
+What happens if we have multiple columns that are non-numeric and we don't want to remove them all to use the built-in `colMeans()` function? We can make our **own** function!
+
+
+{% highlight r %}
+columnmean <- function (y, remove.NA = TRUE) {
+ nc <- ncol(y)
+ means <- vector("numeric", length=0)
+ for (i in 1:nc) {
+ means[i] <- mean(y[,i], na.rm = remove.NA)
+ }
+ means
+}
+
+columnmean(airQuality)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## Warning in mean.default(y[, i], na.rm = remove.NA): argument is not numeric
+## or logical: returning NA
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 42.129310 185.931507 9.957516 77.882353 NA 15.803922
+## [7] 0.916601
+{% endhighlight %}
+
+Cool right? Now let's try a function with an if/else expression.
+
+
+{% highlight r %}
+
+above80 <- function(y) {
+ y <- na.omit(y)
+ nob <- length(y)
+ high <- vector("numeric", length=0)
+ low <- vector("numeric", length=0)
+ for (i in 1:nob) {
+ if (y[i] > 80) {
+ high <- append(high, y[i])
+ }
+ else {
+ low <- append(low, y[i])
+ }
+ }
+ list(high,low)
+}
+
+above80(airQuality$Temp)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [[1]]
+## [1] 81 84 85 82 87 90 87 93 92 82 83 84 85 81 84 83 83 88 92 92 89 82 81
+## [24] 91 81 82 84 87 85 81 82 86 85 82 86 88 86 83 81 81 81 82 86 85 87 89
+## [47] 90 90 92 86 86 82 81 86 88 97 94 96 94 91 92 93 93 87 84 81 82 81
+##
+## [[2]]
+## [1] 67 72 74 62 56 66 65 59 61 69 74 69 66 68 58 64 66 57 68 62 59 73 61
+## [24] 61 57 58 57 67 79 76 78 74 67 79 80 79 77 72 65 73 76 77 76 76 76 75
+## [47] 78 73 80 77 73 80 74 80 79 77 79 76 78 78 77 72 75 79 80 78 75 73 76
+## [70] 77 71 71 78 67 76 68 64 71 69 63 70 77 75 76 68
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+highlow <- above80(airQuality$Temp)
+
+high <- highlow[[1]]
+
+low <- highlow[[2]]
+{% endhighlight %}
+
+# Loop Functions #
+
+The last thing for today are the very useful built-in loop functions. These functions have the word `apply` in them.
+
+- `lapply`: loop over a list and evaluate a function on each element
+- `sapply`: same as `lapply` but it tries to simplify the result
+- `apply`: apply a function over the margins of an array
+- `tapply`: apply a function over subsets of a vector (`lapply()` used with `split()` does the same thing)
+- `mapply`: a multivariate version of `lapply`
+
+# lapply() #
+
+Remember the function we made? It can be replaced with an `lapply()` loop.
+
+{% highlight r %}
+lapply(airQuality, mean, na.rm = TRUE)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## Warning in mean.default(X[[i]], ...): argument is not numeric or logical:
+## returning NA
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## $Ozone
+## [1] 42.12931
+##
+## $Solar.R
+## [1] 185.9315
+##
+## $Wind
+## [1] 9.957516
+##
+## $Temp
+## [1] 77.88235
+##
+## $Month
+## [1] NA
+##
+## $Day
+## [1] 15.80392
+##
+## $Random
+## [1] 0.916601
+{% endhighlight %}
+
+# sapply() #
+
+Alternatively we can try `sapply()`. This will return a named numeric vector (simpified from a list)
+
+{% highlight r %}
+sapply(airQuality, mean, na.rm = TRUE)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## Warning in mean.default(X[[i]], ...): argument is not numeric or logical:
+## returning NA
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## Ozone Solar.R Wind Temp Month Day
+## 42.129310 185.931507 9.957516 77.882353 NA 15.803922
+## Random
+## 0.916601
+{% endhighlight %}
+
+# apply() #
+
+`apply()` first coerces your dataframe to a matrix, which means all the columns must have the same type. Because our dataframe does not meet this requirement, I'll be taking a subset of the data. However normally this function would be used with matrices.
+
+
+{% highlight r %}
+apply(airQuality[,c(-5)], 2, sd, na.rm = TRUE)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## Ozone Solar.R Wind Temp Day Random
+## 32.987885 90.058422 3.523001 9.465270 8.864520 2.091644
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+apply(airQuality[,c(-5)], 1, sd, na.rm = TRUE)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 72.89058 46.94296 58.69988 121.77277 25.62174 25.75497 115.47911
+## [8] 38.64267 21.34867 81.09362 30.02192 98.62413 112.61584 105.66532
+## [15] 27.26847 129.50176 116.47159 31.26497 123.08330 23.93098 21.81966
+## [22] 123.51997 23.04112 33.19412 26.94703 110.01967 25.26733 22.88824
+## [29] 92.82220 84.03591 104.86704 122.41928 122.31465 102.42537 79.76670
+## [36] 94.18712 111.56434 51.01858 116.07814 110.38572 123.47021 109.27081
+## [43] 105.86196 58.30500 140.07391 135.54122 72.33150 106.82007 23.63969
+## [50] 46.01597 53.22244 63.11980 33.84056 41.67048 103.81005 55.83172
+## [57] 53.47395 29.03317 42.86913 29.24985 57.85399 106.70907 95.40798
+## [64] 91.31892 48.18947 67.71641 120.77618 105.84699 102.34370 104.07476
+## [71] 67.97639 60.74067 101.58831 66.48642 121.75326 30.67241 98.43243
+## [78] 104.27405 107.64244 72.14305 81.66779 26.80088 107.47906 122.58368
+## [85] 109.45345 84.18191 35.62429 36.17203 79.75871 103.09283 93.26686
+## [92] 94.07138 38.88395 29.95196 38.00552 43.19716 35.16671 41.00030
+## [99] 99.41880 87.31363 81.25121 94.66409 59.49559 73.33740 104.78695
+## [106] 59.10313 35.31902 32.01702 31.20583 45.29166 91.96314 70.91207
+## [113] 98.68951 25.94546 105.50772 78.26531 97.84691 80.28066 64.05905
+## [120] 75.83915 86.56595 88.41150 70.34087 68.32943 77.89078 72.86685
+## [127] 75.04048 43.14471 39.50083 97.28408 85.08832 88.34328 99.40968
+## [134] 89.14843 99.03010 90.96073 25.09182 44.32411 89.59789 84.73321
+## [141] 27.09205 90.80568 77.70208 91.01537 25.24025 52.22038 26.51072
+## [148] 21.49442 72.38703 58.93778 72.25072 50.24947 83.25093
+{% endhighlight %}
+
+# mapply #
+
+`mapply` is a multivariate apply (like `lapply`) that applies a function over a set of arguments.
+
+{% highlight r %}
+mapply(range, airQuality[,c(1,3,6)], na.rm = TRUE)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## Ozone Wind Day
+## [1,] 1 1.7 1
+## [2,] 168 20.7 31
+{% endhighlight %}
+
+# tapply #
+
+`tapply` is used to apply a function over subsets of a vector. It takes two arguements: a vector to apply to function on and a factor variable that subsets the vector. It will simplify the result, like sapply.
+
+
+{% highlight r %}
+tapply(airQuality$Ozone, airQuality$Month, range, na.rm = TRUE)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## $Aug
+## [1] 9 168
+##
+## $Jul
+## [1] 7 135
+##
+## $Jun
+## [1] 12 71
+##
+## $May
+## [1] 1 115
+##
+## $Sep
+## [1] 7 96
+{% endhighlight %}
+
diff --git a/lessons/rintro/slides.md b/lessons/rintro/slides.md
new file mode 100644
index 0000000..22b8b96
--- /dev/null
+++ b/lessons/rintro/slides.md
@@ -0,0 +1,469 @@
+---
+title: "Introduction to R"
+author: Sarah Meister
+date: 2015-11-02
+layout: page
+sidebar: false
+tag:
+ - Lessons
+ - Slides
+ - IntroR
+ - R
+categories:
+ - Lessons
+ - IntroR
+ - R
+---
+
+# Welcome to our R introduction workshop #
+
+## Purpose: ##
+
+To introduce you to the R language and common data manipulations used in R
+
+# What is R and why do I need to learn it? #
+
+R is an object-oritented programming language for statistical computing and graphics.
+
+Benefits of R:
+
+1. Easily extendible through **FREE** R packages
+2. Active community (lots on online help)
+3. Able to produce publication-quality graphs, and dynamic and interactive graphs with additional packages (did I mention they were **FREE**??)
+4. For computationally intensive tasks, R objects can be manipulated with C, C++, Java, .NET or Python code
+5. Has it's own flavor of Markdown, making R code easily documented and shareable (which you will be learning in another lesson)
+
+## Learning objectives ##
+
+After this workshop, our expectation is that you will be able to:
+
+1. Understand the "grammar" of the R language
+2. Import and export data into R
+3. View data in R and perform basic calculations & manipulations
+4. Make and use a basic R function
+5. Understand and use the useful built-in loop functions
+
+# Materials for this lesson: #
+
+* [Slides](slides/)
+* [Cheatsheet](cheatsheet/)
+* [Assignment](assignment/)
+* [Assignment Answers](answer)
+
+Other resources can be found [here](../resources/)
+
+# The R language #
+
+R is an object-oriented programing language, meaning everything we encounter in R is an object.
+
+R has a 5 basic classes of objects:
+
+* Character
+* Numeric
+* Integer
+* Complex
+* Logical
+
+R has 5 different types of objects:
+
+* Vectors
+* Lists
+* Matrices
+* Factors
+* Dataframes
+
+Thus you could have a logical vector, a character dataframe, a numeric matrix, etc.
+
+# Vectors #
+
+The most basic object is a vector. A vector is a sequence of objects of the same class. We can make vectors using the `c()` function.
+
+
+{% highlight r %}
+a <- c(0.4, 1.2) # numeric
+b <- c(TRUE, FALSE) # logical
+c <- c("a","b","c") # character
+d <- c(1L, 2L) # integer
+e <- 1:10 # integer
+f <- c(1+0i, 2+4i) # complex
+{% endhighlight %}
+
+Empty vectors can be made using the `vector()` function
+
+
+{% highlight r %}
+x <- vector("numeric", length=10)
+x
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 0 0 0 0 0 0 0 0 0 0
+{% endhighlight %}
+
+# Lists #
+
+A list is a special type of vector that can contain objects of different classes. For instance, we can have a list with a numeric dataframe as one object within the list, and a string vector as another.
+
+
+{% highlight r %}
+x <- list(1,"a", TRUE, 1+4i)
+x
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [[1]]
+## [1] 1
+##
+## [[2]]
+## [1] "a"
+##
+## [[3]]
+## [1] TRUE
+##
+## [[4]]
+## [1] 1+4i
+{% endhighlight %}
+
+The elements of a list are seperated by double brackets.
+
+# Matrices #
+
+Matrices are vectors with a dimension attribute. The `dim` attribute is an integer vector of length = 2 (nrow, ncol). Matrix multiplication is done with the %*% operator
+
+
+{% highlight r %}
+x <- matrix(1:6, nrow=2, ncol=3)
+x
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [,1] [,2] [,3]
+## [1,] 1 3 5
+## [2,] 2 4 6
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+dim(x)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 2 3
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+attributes(x)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## $dim
+## [1] 2 3
+{% endhighlight %}
+
+Matrices are constructed column-wise, so entries start at the upper left corner and run down the columns.
+
+Matrices can be created directly from vectors by adding a dimension value
+
+
+{% highlight r %}
+x <- 1:10
+
+x
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 1 2 3 4 5 6 7 8 9 10
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+dim(x) <- c(2,5)
+
+x
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [,1] [,2] [,3] [,4] [,5]
+## [1,] 1 3 5 7 9
+## [2,] 2 4 6 8 10
+{% endhighlight %}
+
+We can also create matrices by column-binding or row-binding with the functions `cbind()` and `rbind()`
+
+
+{% highlight r %}
+x <- 1:3
+y <- 10:12
+cbind(x,y)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## x y
+## [1,] 1 10
+## [2,] 2 11
+## [3,] 3 12
+{% endhighlight %}
+
+
+
+{% highlight r %}
+rbind (x, y)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [,1] [,2] [,3]
+## x 1 2 3
+## y 10 11 12
+{% endhighlight %}
+
+# Factors #
+
+Factors are used to represent catagorical data. They can be ordered or unordered.
+
+* They are like an integer vector where each integer has a label.
+* Factors are more descriptive than integers e.g. "Male" and "Female" vs. 1 and 2
+* Factors are treated specially by modelling functions like `lm()` and `glm()`
+
+
+{% highlight r %}
+x <- factor(c("yes", "yes", "no", "no", "yes"))
+x
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] yes yes no no yes
+## Levels: no yes
+{% endhighlight %}
+
+
+
+{% highlight r %}
+table (x)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## x
+## no yes
+## 2 3
+{% endhighlight %}
+
+
+
+{% highlight r %}
+unclass(x)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 2 2 1 1 2
+## attr(,"levels")
+## [1] "no" "yes"
+{% endhighlight %}
+
+# Dataframe #
+
+Dataframes are used in R to store tabular data. Dataframes are like matrices (meaning they have rows and columns) that can store different classes of objects in each column (e.g. numeric, character, logical, etc.).
+
+Depending on the source of the data, dataframes can be made using the `read.table()`, `read.csv()` or `data.frame()` functions.
+
+
+{% highlight r %}
+(x <- data.frame(foo = 1:4, bar = c(T,T,F,F)))
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## foo bar
+## 1 1 TRUE
+## 2 2 TRUE
+## 3 3 FALSE
+## 4 4 FALSE
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+nrow(x)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 4
+{% endhighlight %}
+
+
+
+{% highlight r %}
+ncol(x)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] 2
+{% endhighlight %}
+
+
+# Attributes #
+
+R objects can have attributes, such as:
+
+* names
+* classes
+* lengths
+* dimensions (matrices/dataframes)
+* user defined attributes, such as metadata
+* dataframes have a special attribute called `row.names`
+
+Attributes can be modified with the `attributes()` function, among others.
+
+For example, modifiying the `names` of a vector can make your code more readable.
+
+{% highlight r %}
+x <- 1:3
+names(x)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## NULL
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+names(x) <- c("alpha", "beta", "gamma")
+
+x
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## alpha beta gamma
+## 1 2 3
+{% endhighlight %}
+
+
+
+{% highlight r %}
+names(x)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## [1] "alpha" "beta" "gamma"
+{% endhighlight %}
+
+
+
+{% highlight r %}
+str(x)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## Named int [1:3] 1 2 3
+## - attr(*, "names")= chr [1:3] "alpha" "beta" "gamma"
+{% endhighlight %}
+
+# Now let's code! #
+
+# Starting R #
+
+We will be using R studio for this course as it's a user-friendly GUI for R. You can also access R from Terminal (Mac) or Command Line (Windows), if you please.
+
+Download the airQuality.csv file from our [Github repo](https://github.com/codeasmanuscript/workshops/tree/master/lessons/rintro) and import it into R using the `read.csv` function
+
+# Loop Functions #
+
+R has some very useful built-in loop functions. These functions have the word `apply` in them.
+
+- `lapply`: loop over a list and evaluate a function on each element
+- `sapply`: same as `lapply` but it tries to simplify the result
+- `apply`: apply a function over the margins of an array
+- `tapply`: apply a function over subsets of a vector (`lapply()` used with `split()` does the same thing)
+- `mapply`: a multivariate version of `lapply`
+
+# lapply() #
+
+Remember the function we made? It can be replaced with an `lapply()` loop.
+
+{% highlight r %}
+lapply(airQuality, mean, na.rm = TRUE)
+{% endhighlight %}
+
+# sapply() #
+
+Alternatively we can try `sapply()`. This will return a named numeric vector (simpified from a list)
+
+{% highlight r %}
+sapply(airQuality, mean, na.rm = TRUE)
+{% endhighlight %}
+
+# apply() #
+
+`apply()` first coerces your dataframe to an array which means all the columns must have the same type. Because our dataframe does not meet this requirement, I'll be taking a subset of the data. Normally this function would only be used with matrices.
+
+
+{% highlight r %}
+apply(airQuality[,c(-5)], 2, sd, na.rm = TRUE)
+
+apply(airQuality[,c(-5)], 1, sd, na.rm = TRUE)
+{% endhighlight %}
+
+# mapply #
+
+`mapply` is a multivariate apply (like `lapply`) that applies a function over a set of arguments.
+
+{% highlight r %}
+mapply(range, airQuality[,c(1,3,6)], na.rm = TRUE)
+{% endhighlight %}
+
+# tapply #
+
+`tapply` is used to apply a function over subsets of a vector. It takes two arguements: a vector to apply to function on and a factor variable that subsets the vector. It will simplify the result, like sapply.
+
+
+{% highlight r %}
+tapply(airQuality$Ozone, airQuality$Month, range, na.rm = TRUE)
+
+## applying the range function on the Ozone column by the Month column.
+{% endhighlight %}
diff --git a/lessons/rmarkdown/assignment.md b/lessons/rmarkdown/assignment.md
new file mode 100644
index 0000000..9bb97b3
--- /dev/null
+++ b/lessons/rmarkdown/assignment.md
@@ -0,0 +1,48 @@
+---
+title: "Assignment: R Markdown"
+published: true
+author:
+ - Luke W. Johnston
+date: 2015-10
+layout: page
+sidebar: false
+tag:
+ - Lessons
+ - Assignment
+ - Rmarkdown
+ - R
+categories:
+ - Lessons
+ - Rmarkdown
+ - R
+output:
+ md_document:
+ variant: markdown_github
+---
+
+## Challenges: Try these out yourself!
+
+Try to progressively add these challenges into an `.Rmd` file. First, create a
+template R Markdown file from RStudio. Push the final version to the GitHub
+repo, so we can look them over and make suggestions.
+
+1. Create section 'Brief description' describing your assigned dataset. Use
+inline R code to describe some basic statistics of the variables (eg. means,
+standard deviation, number of variables, and number of rows).
+
+2. Create a new section 'Plots' for some plots of some of the variables in the
+dataset. Include the code from the `plotting` workshop assignment into the
+`.Rmd` file, replacing the old dataset with the new dataset name (including the
+variable names too!). Include a figure caption (using the YAML option and the
+code chunk option) and increase the DPI of the figure. Use `captioner` to create
+the figure caption.
+
+3. Create another section 'Tables' for some tables. Include the R code from
+`wrangling` workshop assignment into the `.Rmd` file. Replace the old dataset
+with the new dataset name, as well as the variable names. Convert the `%>%` pipe
+chain to a table using `pander`. Use `captioner` for the table captions.
+
+4. Generate both a Word file and an HTML file at the same time, using the YAML
+metadata and the `render` command. Change the theme of the HTML document and
+number the sections, and include a table of contents (toc) into both the Word
+and HTML documents.
diff --git a/lessons/rmarkdown/cheatsheet.md b/lessons/rmarkdown/cheatsheet.md
new file mode 100644
index 0000000..73eec8a
--- /dev/null
+++ b/lessons/rmarkdown/cheatsheet.md
@@ -0,0 +1,139 @@
+---
+title: "Cheatsheet: R Markdown"
+published: true
+author:
+ - Luke W. Johnston
+date: 2015-10
+layout: page
+sidebar: false
+tag:
+ - Lessons
+ - Cheatsheet
+ - Rmarkdown
+ - R
+categories:
+ - Lessons
+ - Rmarkdown
+ - R
+output:
+ md_document:
+ toc: true
+ variant: markdown_github
+---
+
+
+
+Want a streamlined and efficient workflow where you wrangle your data, analyze
+it, plot it or make a table of the results, and put it into your manuscript or
+thesis all in one go? Well, you are lucky! R Markdown does exactly take, by
+making you more productive and efficient! Use this cheatsheet as a reference to
+learning how to use R Markdown, or better yet, check out the [R resources list](../resources/)
+for a printable cheatsheet (developed by RStudio).
+
+# R Markdown commands: Some useful or common ones #
+
+## YAML metadata ##
+
+> Most R Markdown files will contain YAML metadata for options on how to
+generate the output document. These options are used by R Markdown to tell
+pandoc (the conversion tool) how to create the document.
+
+> Example YAML options:
+
+ ---
+ title: 'Title here'
+ author: 'Author here'
+ date: 2015-10-10
+ bibliography: /path/to/file
+ output:
+ html_document:
+ toc: true
+ number_section: true
+ fig_caption: true
+ word_document:
+ toc: true
+ fig_caption: true
+ ---
+
+## R code chunk options ##
+
+> Code chunks in `.Rmd` files are started with three backticks, r, the chunk
+name, and the options, which would look like ` ```{r chunkName, echo=FALSE}``` `.
+Code chunks allow R to run the code inside and print the R output onto the
+HTML/Word/PDF document. The chunk options customize the output as specified.
+
+> Example options:
+
+ eval=TRUE
+ echo=TRUE
+ message=TRUE
+ error=TRUE
+ warning=TRUE
+ results=('markup', 'asis', 'hide')
+ fig.height=7
+ fig.width=7
+ dpi=90
+ fig.cap='Caption title'
+
+## `pander` ##
+
+> The `pander` function in the `pander` package can create a markdown table from
+an R object. This lets you easily change those pesky tables, no longer needing
+to copy and paste into the table! You will need to specific the code chunk
+option `results='asis'` to make R print the table correctly. `pander` tables
+can be highly customized. Please see the [resources](../resources/) or go
+directly to the [`pander` website](http://rapporter.github.io/pander/) for a
+detailed reference to completely customizing tables.
+
+> Example code:
+
+ library(pander)
+ pander(head(cars))
+ ## Or print linear regression output:
+ pander(lm(cars))
+ ## Or t-test
+ pander(t.test(cars))
+ ## Include a caption for the next table:
+ set.caption('Caption title here')
+ ## Table of correlations coefficients
+ pander(cor(cars))
+ ## Or directly include the caption in the command:
+ pander(cor(cars), caption = 'Caption title here')
+
+## `captioner` ##
+
+> Depending on the output format, there will likely not be a number assigned to
+each table or figure. If your table or figure changes frequently or based on
+reviewer comments, you may want to assign a label to the table or figure and
+have it change the numbering for you. These can all be fixed by using
+`captioner` from the `captioner` package.
+
+> Example code:
+
+
+{% highlight r %}
+ library(captioner)
+ tabNums <- captioner(prefix = 'Table')
+ tabNums('tab1', 'Caption for table 1')
+ ## cite in text using `r tabNums('tab1', display = 'cite')`
+ pander(head(cars), caption = tabNums('tab1'))
+
+ ## For figures...
+ figNums <- captioner(prefix = 'Figure')
+ figNums('fig1', 'Caption for figure 1')
+ ## cite in text using `r figNums('fig1', display = 'cite')`
+ ## Include the caption for the figure in the code chunk
+ ## using fig.cap=`r figNums('fig1')`
+{% endhighlight %}
+
+## `render` ##
+
+> This is the command to convert the `.Rmd` or `.R` file into other document
+formats, including Word, HTML, or PDF. Conversion to PDF requires a working
+LaTeX distribution (see [here](https://www.latex-project.org/) for help).
+
+> Example code:
+
+ library(rmarkdown)
+ ## Convert to word and html files.
+ render('file.Rmd', c('word_document', 'html_document'))
diff --git a/lessons/rmarkdown/intro.md b/lessons/rmarkdown/intro.md
new file mode 100644
index 0000000..6212086
--- /dev/null
+++ b/lessons/rmarkdown/intro.md
@@ -0,0 +1,288 @@
+---
+title: "Introduction: Report generation using R Markdown"
+published: true
+author:
+ - Luke W. Johnston
+date: 2015-10
+layout: page
+sidebar: false
+tag:
+ - Lessons
+ - Introduction
+ - Rmarkdown
+ - R
+categories:
+ - Lessons
+ - Rmarkdown
+ - R
+permalink: lessons/rmarkdown/
+output:
+ html_document:
+ toc: true
+ md_document:
+ variant: markdown_github
+ toc: true
+---
+
+A recent feature to using R is the incredibly powerful and incredibly useful
+package [`rmarkdown`](http://rmarkdown.rstudio.com/) (and by extension the
+package [`knitr`](http://yihui.name/knitr/)). R Markdown allows you to
+dynamically generate a report (could be a manuscript or your thesis!) where R
+automatically inserts tables, figures, numbers, and citations into your document
+so that you never again have to copy and paste anything! You can create Word,
+HTML, or PDF files from your `.Rmd` (or R Markdown) file, letting you
+collaborate with the least tech-savvy individuals. It also dramatically cuts
+down on your time writing your manuscript or thesis because as you write it up
+you are also putting in the R code to create everything that will be in the
+report. In one step you go for analysis to report! No more worries about fixing
+up your report after a co-author suggests to re-do a part of the analysis!
+
+# Learning objectives:
+
+1. Write in markdown
+2. Use YAML and R code chunk options
+3. Create tables and figures
+4. Ultimately, generate a dynamic report in either Word or HTML.
+
+# Materials for this lesson:
+
+* [Slides](slides/)
+* [Cheatsheet](cheatsheet/)
+* [Assignment](assignment/)
+
+More resources on R Markdown can be found [here](../resources/).
+
+# Generating a dynamic report!
+
+## YAML metadata
+
+This is the part that tells R how to generate the document.
+[YAML](https://en.wikipedia.org/wiki/YAML) is a human-readable data-encoding
+format. What that means is it makes it so you can easily type up commands that
+both you and the computer will understand. A typical YAML metadata looks like:
+
+```
+---
+title: "Introducing R Markdown"
+published: true
+author: "Luke Johnston"
+date: "July 23, 2015"
+output: html_document
+---
+```
+
+Note the starting and ending `---` 'tags', which tell R that this is a YAML
+block.
+
+## Markdown syntax
+
+Markdown is called a 'markup language' that allows you to convert to a vast
+number of file formats (html, word, pdf, etc). It's a 'language' because you use
+specific characters or symbols to represent something else in another file
+format after conversion. For instance, see below:
+
+```
+# This is a header
+
+## This is a second-level header
+
+Let's show some **bolding** and *italics*. Or why not show a link to [Google](http://google.ca). Some times you may need to show `code` snippets. Or a code block, either using tab or three backticks (```):
+
+ code <- here
+
+Sometimes we need a list:
+
+- Item 1
+- Item 2
+
+Or a numbered list:
+
+1. Item 1
+2. Item 2
+
+This is a citation @Joe2005 (not converted). Or an image (not converted):
+
+
+
+```
+
+## R Markdown
+
+R Markdown is Markdown and R combined! So you get the simplicity of Markdown
+with the power of R! Within the R Markdown document, you can insert R code
+chunks to input and create the output into the document. They look like this:
+
+
+ {% highlight r %}
+ testCode <- 1:10
+ print(testCode)
+ {% endhighlight %}
+
+
+
+ {% highlight text %}
+ ## [1] 1 2 3 4 5 6 7 8 9 10
+ {% endhighlight %}
+
+You'll notice that the code chunk printed off 1 to 10 and put it into the
+document. I didn't do that, R did it for me!
+
+The code chunks have several options that allow you to customize how you want R
+to treat the chunk. Most options require either a `TRUE` or `FALSE` as the
+setting. Two commonly used options include:
+
+- `eval`: Whether or not R runs the code chunk
+- `echo`: Whether the code chunk is kept in the converted format (eg. Word or
+HTML)
+
+There are also options `message`, `warning`, `error`.
+
+You can also include inline R code snippets that pastes the output directly into
+the paragraph. Use the markdown syntax for inline code, except with an r
+infront like ` testing `, which creates testing.
+
+## Creating tables using R code
+
+Within Markdown is the ability to create tables the convert into the other
+document formats, such as Word or HTML. Using the `pander()` command from the
+`pander` package, you can create these Markdown tables of an R object. In
+addition to the `pander()` command, there is an extra step to take to get the
+object to print: including a `results = 'asis'` option in the code chunk. Here
+is an example:
+
+
+{% highlight r %}
+library(pander)
+pander(head(cars), caption = 'First few rows of the cars dataset.',
+ style = 'rmarkdown')
+{% endhighlight %}
+
+
+
+| speed | dist |
+|:-------:|:------:|
+| 4 | 2 |
+| 4 | 10 |
+| 7 | 4 |
+| 7 | 22 |
+| 8 | 16 |
+| 9 | 10 |
+
+Table: First few rows of the cars dataset.
+
+## Figures in the markdown file
+
+Just like you can get R to create a table, you can also get R to create a
+figure. There are several chunk options for creating figures, including `fig.height`, `fig.width`, `dpi`, and `fig.cap`.
+
+
+{% highlight r %}
+plot(cars)
+{% endhighlight %}
+
+
+
+## Bibliography
+
+If you want to cite articles or references, use the `@` tag with the author key
+(eg. `@Smith1995`). In addition to the `@` tag, you need to include the
+following YAML option to the YAML header, with the location of the bibliography
+file on your computer.
+
+```
+bibliography: /path/to/file
+```
+
+## Figure and table labels
+
+> *Note: The below labels don't work with conversion on the website. See [this
+link](intro.html) for the HTML converted version that chows these labels
+working.*
+
+Often you want to include figure or caption labels that you can cite easily in
+your document. So, let's create two labels for a table and a figure using the
+`captioner` command from the `captioner` package. First we set the prefix for
+the caption:
+
+
+{% highlight r %}
+library(captioner)
+figNums <- captioner(prefix = 'Figure')
+tabNums <- captioner(prefix = 'Table')
+{% endhighlight %}
+
+Then, we store the caption label and caption title (including the `results='hide'`
+code chunk option, as these commands will print them to the output):
+
+
+{% highlight r %}
+figNums(name = 'figCars', caption = 'First few rows of the cars dataset.',
+ display = FALSE)
+tabNums(name = 'tabCars', caption = 'Scatterplot of speed and distance.',
+ display = FALSE)
+{% endhighlight %}
+
+Now we can cite them in-text, using the ` Figure 1 `,
+which then looks like Figure 1. And with the
+plot, include this option in the plot code chunk ` Figure 1: First few rows of the cars dataset. `:
+
+
+{% highlight r %}
+plot(cars)
+{% endhighlight %}
+
+
+
+Or with a table using ` Table 1 ` to show
+Table 1.
+
+
+{% highlight r %}
+set.caption(tabNums('tabCars'))
+pander(head(cars), style = 'rmarkdown')
+{% endhighlight %}
+
+
+
+| speed | dist |
+|:-------:|:------:|
+| 4 | 2 |
+| 4 | 10 |
+| 7 | 4 |
+| 7 | 22 |
+| 8 | 16 |
+| 9 | 10 |
+
+Table: Table 1: Scatterplot of speed and distance.
+
+## Generating this `.Rmd` document and sharing with others
+
+If you download the [source `.Rmd` version of this document](https://github.com/codeasmanuscript/workshops/tree/master/lessons/rmarkdown/intro.Rmd),
+you can recreate the web version and the HTML version by using these commands
+below (note the `eval=FALSE` in the code chunk):
+
+
+{% highlight r %}
+library(rmarkdown)
+## html
+render('intro.Rmd', 'html_document')
+## md version on the website
+render('intro.Rmd', 'md_document')
+{% endhighlight %}
+
+R Markdown can also make it very easy to collaborate with people who may not be
+as tech-savvy and who don't know much outside of Microsoft Word by converting
+all your `.Rmd` files into Word! Try it out:
+
+
+{% highlight r %}
+render('intro.Rmd', 'word_document')
+{% endhighlight %}
+
+Using R Markdown can help with making your code easier to understand. That's
+because as you type out your code, you are also typing explanations and reasons
+in markdown. You can then easily create documents, manuscripts, theses, and
+other file types that others (and your future self) can read to understand your
+code and your analysis better. Combine R and R Markdown with Git and GitHub and
+you have a powerful tool for making your work better, more scientifically
+rigorous and transparent, and share-able so others can use your work easily!
diff --git a/lessons/rmarkdown/slides.md b/lessons/rmarkdown/slides.md
new file mode 100644
index 0000000..568c950
--- /dev/null
+++ b/lessons/rmarkdown/slides.md
@@ -0,0 +1,37 @@
+---
+title: "Dynamic report generation using R Markdown and knitr"
+published: true
+author: Luke W. Johnston
+date: 2015-10
+layout: page
+sidebar: false
+tag:
+ - Lessons
+ - Slides
+ - Rmarkdown
+ - R
+categories:
+ - Lessons
+ - Rmarkdown
+ - R
+output: slidy_presentation
+---
+
+## Learning expectations ##
+
+- How to write reproducible reports
+- How to write in markdown
+- How to set the report options
+- How to make tables and figures in the report
+
+## 5 main concepts: ##
+
+- **Markdown syntax**: `#` or `##`, `*`, `**`, `![]()`, ` ``` `, \`, `>`, `-`,
+`@`, `[]()`
+- **YAML**: output, `word_document`, `html_document`, bibliography
+- **Options**: echo, eval, message, warning, error, fig.height, fig.width,
+results, fig.cap
+- **Tables and figures**: pander, kable, ggplot, qplot, captioner
+- **Generate the report**: knitr, render, 'Knit HTML' or 'Knit Word' button
+(RStudio)
+
diff --git a/lessons/rplotting/assignment.md b/lessons/rplotting/assignment.md
new file mode 100644
index 0000000..2ac5889
--- /dev/null
+++ b/lessons/rplotting/assignment.md
@@ -0,0 +1,23 @@
+---
+title: "Assignment: ..."
+author:
+ -
+date: YYYY-MM-DD
+fontsize: 12pt
+geometry: margin=1in
+papersize: letterpaper
+layout: page
+sidebar: false
+tag:
+ - Lessons
+ - Assignment
+ - (lesson topic)
+categories:
+ - Lessons
+ - (lesson topic)
+---
+
+# List of potential exercises here: #
+
+* List item 1
+* List item 2
diff --git a/lessons/rplotting/cheatsheet.md b/lessons/rplotting/cheatsheet.md
new file mode 100644
index 0000000..ab1c6a3
--- /dev/null
+++ b/lessons/rplotting/cheatsheet.md
@@ -0,0 +1,39 @@
+---
+title: "Cheatsheet: (topic)"
+author:
+ -
+date: YYYY-MM-DD
+fontsize: 12pt
+geometry: margin=1in
+papersize: letterpaper
+layout: page
+sidebar: false
+tag:
+ - Lessons
+ - Cheatsheet
+ - (lesson topic)
+categories:
+ - Lessons
+ - (lesson topic)
+---
+
+Brief description/intro
+
+# (language) commands: Some useful or common ones #
+
+## `command` ##
+
+> Description
+
+> Example code:
+
+ example code (note the tab/4 spaces indent)
+
+## `command` ##
+
+> Description
+
+> Example code:
+
+ example code (note the tab/4 spaces indent)
+
diff --git a/lessons/rplotting/plottinginR.md b/lessons/rplotting/plottinginR.md
new file mode 100644
index 0000000..4ed8a5a
--- /dev/null
+++ b/lessons/rplotting/plottinginR.md
@@ -0,0 +1,156 @@
+---
+title: "Plotting in R"
+author: "Sarah Meister"
+date: 2015-10-15
+fontsize: 12pt
+geometry: margin=1in
+papersize: letterpaper
+layout: page
+sidebar: false
+output: slidy_presentation
+---
+
+# Base plotting systems #
+
+The base plotting system in R is the `plot()` function.
+
+
+{% highlight r %}
+library(datasets)
+data(cars)
+with(cars, plot(speed, dist))
+{% endhighlight %}
+
+
+
+# the Lattice system #
+
+This is the second base plotting system in R. Plots are created with a single call function. Margins and spacing are set automatically because the entire plot is specified at once. It is good at putting many plots on a screen and thus you can see things like how y changes with x over z.
+
+
+{% highlight r %}
+library (lattice)
+state <- data.frame(state.x77, region = state.region)
+xyplot(Life.Exp ~ Income | region, data=state, layout = c(4,1))
+{% endhighlight %}
+
+
+
+# ggplot2 #
+
+The ggplot2 package is an R package that uses the "Grammar of Graphics" to put together different aspects of data visualization to build quality graphics. It is a very popular package.
+
+
+ {% highlight r %}
+ install.packages("ggplot2")
+ {% endhighlight %}
+
+
+
+ {% highlight text %}
+ ## Installing package into '/home/luke/R/x86_64-pc-linux-gnu-library/3.2'
+ ## (as 'lib' is unspecified)
+ {% endhighlight %}
+
+
+
+ {% highlight text %}
+ ## Error in contrib.url(repos, type): trying to use CRAN without setting a mirror
+ {% endhighlight %}
+
+
+
+ {% highlight r %}
+
+ library(ggplot2)
+ {% endhighlight %}
+
+
+
+ {% highlight text %}
+ ## Loading required package: methods
+ {% endhighlight %}
+
+ # the Basics #
+
+ `qplot()` is the basic plotting function in `ggplot2` and plots are made up of aesthetics (size, shape, color, etc.) and geoms (points, lines, etc.).
+
+ `ggplot()` is the core function and can do things that `qplot` cannot.
+
+
+{% highlight r %}
+str(mpg)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## 'data.frame': 234 obs. of 11 variables:
+## $ manufacturer: Factor w/ 15 levels "audi","chevrolet",..: 1 1 1 1 1 1 1 1 1 1 ...
+## $ model : Factor w/ 38 levels "4runner 4wd",..: 2 2 2 2 2 2 2 3 3 3 ...
+## $ displ : num 1.8 1.8 2 2 2.8 2.8 3.1 1.8 1.8 2 ...
+## $ year : int 1999 1999 2008 2008 1999 1999 2008 1999 1999 2008 ...
+## $ cyl : int 4 4 4 4 6 6 6 4 4 4 ...
+## $ trans : Factor w/ 10 levels "auto(av)","auto(l3)",..: 4 9 10 1 4 9 1 9 4 10 ...
+## $ drv : Factor w/ 3 levels "4","f","r": 2 2 2 2 2 2 2 1 1 1 ...
+## $ cty : int 18 21 20 21 16 18 18 18 16 20 ...
+## $ hwy : int 29 29 31 30 26 26 27 26 25 28 ...
+## $ fl : Factor w/ 5 levels "c","d","e","p",..: 4 4 4 4 4 4 4 4 4 4 ...
+## $ class : Factor w/ 7 levels "2seater","compact",..: 2 2 2 2 2 2 2 2 2 2 ...
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+qplot(displ, hwy, data = mpg)
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+qplot(displ, hwy, data = mpg, color=drv)
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+qplot(displ, hwy, data = mpg, geom=c("point", "smooth"))
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## geom_smooth: method="auto" and size of largest group is <1000, so using loess. Use 'method = x' to change the smoothing method.
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+qplot(hwy, data=mpg, fill=drv)
+{% endhighlight %}
+
+
+
+{% highlight text %}
+## stat_bin: binwidth defaulted to range/30. Use 'binwidth = x' to adjust this.
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+qplot(displ, hwy, data=mpg, facets =.~drv)
+{% endhighlight %}
+
+
+
+{% highlight r %}
+
+qplot(hwy, data=mpg, facet=drv~., binwidth=2)
+{% endhighlight %}
+
+
diff --git a/lessons/rplotting/slides.md b/lessons/rplotting/slides.md
new file mode 100644
index 0000000..5bfb8c2
--- /dev/null
+++ b/lessons/rplotting/slides.md
@@ -0,0 +1,103 @@
+---
+title: "...title..."
+author:
+date: YYYY-MM-DD
+layout: page
+sidebar: false
+classoption: xcolor=dvipsnames
+tag:
+ - Lessons
+ - Slides
+ - (lesson topic)
+categories:
+ - Lessons
+ - (lesson topic)
+slide-level: 1
+fontsize: 8pt
+header-includes:
+ - \input{../slideOptions.tex}
+---
+
+# Welcome to our Data-related workshop #
+
+## Purpose: ##
+
+To teach a few tips and tricks for more efficiently managing your
+data, tracking your computer files, understanding appropriate
+analytical approaches, and speeding up the process from code to
+tables.
+
+. . .
+
+## Significance: ##
+
+Topics we cover will help you get more comfortable with data, reduce
+the chance of overlooked errors, and give you more control over your
+work. They are also all important parts of a science movement gaining
+increasing attention -- Reproducible Research.
+
+# Caveat: We aren't here to teach statistics #
+
+Need help with stats? Use these resources!
+
+* U of T Statistical Consulting Services ([click here](http://www.utstat.toronto.edu/wordpress/?page_id=25))
+
+*
+
+*
+
+# Overview of other workshops? #
+
+# Notes and help during this workshop #
+
+Go to this website:
+
+
+
+# Slide title #
+
+Text, some **bolded**, or *italics*
+
+__bold__ or _italics_ also works.
+
+[URL link here](http://link/here.com)
+
+```
+ Code block
+```
+
+Or:
+
+ Code block
+
+List here:
+
+* Item 1
+* Item 2
+
+List, but one-by-one 'animation':
+
+> * Item 1 appears first
+> * Item 2 appears second
+
+Inline `code text`
+
+Image:
+
+
+Footnote[^1]
+
+[^1]: Footnote text
+
+# Live coding #
+
+# Main Exercise #
+
+A pause/transition here (the . . .)
+
+. . .
+
+Numbered list
+
+1. Exercise 1
+2. Exercise 2
diff --git a/lessons/slides.md b/lessons/slides.md
new file mode 100644
index 0000000..44044d2
--- /dev/null
+++ b/lessons/slides.md
@@ -0,0 +1,51 @@
+---
+title: "Welcome to *Code As Manuscript*!"
+published: true
+author:
+ - Luke W. Johnston
+ - Sarah Meister
+date: 2015-10
+layout: page
+sidebar: false
+tag:
+ - Lessons
+ - Slides
+ - Rmarkdown
+ - R
+categories:
+ - Lessons
+ - Rmarkdown
+ - R
+output: slidy_presentation
+---
+
+## Learning expectations ##
+
+The expected goal of the workshops is that you will be able to:
+
+> - Put your research under version control using Git.
+> - Push and pull your git research repository to either
+ [GitHub](https://github.com/) or
+ [BitBucket](https://bitbucket.org/).
+> - Learn the basics of R and functions within R
+> - Produce publication quality plots
+> - Quickly wrangle your data into an analyzable format
+> - Reproducibly incorporate R code into your manuscript or thesis to
+ instantly add results and plots (no more copy and paste)
+
+## Schedule ##
+
+Every Monday:
+
+1. Git & GitHub -- Oct. 26th
+2. Basics of R -- Nov. 2nd
+3. Visualization -- Nov. 9th
+4. Data wrangling -- Nov. 16th
+5. Dynamic report generation -- Nov. 23rd
+
+## Site to follow ##
+
+Everything we'll be doing will be through this site:
+
+- http://github.com/codeasmanuscript/practice-2015-10
+