diff --git a/README.md b/README.md
index 7bfef81..ce6ab21 100644
--- a/README.md
+++ b/README.md
@@ -18,13 +18,13 @@ Information given in this course is current as of 30th November 2023.
* [Log in to the frontend server](#log-in-to-the-frontend-server)
* [Basic orientation in your home directory](#basic-orientation-in-your-home-directory)
* [System of software modules](#system-of-software-modules)
-* [Download raw data from NCBI SRA](#download-raw-data-from-ncbi-sra)
+* [Raw reads and quality control](#raw-reads-and-quality-control)
# Introduction
## Aims
-This tutorial, in the brief form of a hands-on course, shows how to process and analyse sequencing data using [MetaCentrum NGI](https://www.metacentrum.cz/en/index.html) (National Grid Infrastructure). Participants will be introduced to the basic usage of MetaCentrum, e.g. how to [log in to the frontend server](https://docs.metacentrum.cz/access/log-in/), how to [manipulate data](https://docs.metacentrum.cz/data/data-within/) properly, how to [start an interactive or batch job](https://docs.metacentrum.cz/basics/jobs/), and how to [display graphical output](https://docs.metacentrum.cz/software/graphical-access/).
+This tutorial, in the brief form of a hands-on course, shows how to process and analyse sequencing data using [MetaCentrum NGI](https://www.metacentrum.cz/en/index.html) (National Grid Infrastructure). Participants will be introduced to the basic usage of MetaCentrum, e.g. how to [log in to the frontend server](https://docs.metacentrum.cz/access/log-in/), how to [manipulate data](https://docs.metacentrum.cz/data/data-within/) properly, how to [start an interactive or batch job](https://docs.metacentrum.cz/computing/run-basic-job/), and how to [display graphical output](https://docs.metacentrum.cz/software/graphical-access/).
In the practical part of the course, we will use publicly available sequencing data (produced by [Illumina](https://www.illumina.com/) and [Oxford Nanopore](https://nanoporetech.com/) platforms) for the _de novo_ hybrid assembly of the bacterial genome - specifically, _Escherichia coli_ strain A0 34/86 (as described in this [paper](https://journals.asm.org/doi/10.1128/mra.00363-23)). Unfortunatelly, processing raw reads, genome assembly and following gene prediction and annotation are processes (especially in the case of larger eukaryotic genomes) that often require time-consuming tuning for optimal parameters and considerable hardware resources.
@@ -54,10 +54,10 @@ To get the full potential of this course, each of the participants should be (or
## Dedicated resources
-As is typical for grid computing, all submitted jobs are sorted into specific [queues](https://docs.metacentrum.cz/advanced/queues-in-meta/) (mainly based on the amount of requested resources). The combination of the required resources and the current infrastructure load determines the delay between the job submission and the start of the calculation. Very demanding jobs can wait in the queue for several days before all the required resources are free. We will use a special queue `MetaSeminar` reserved for this course to avoid this delay. This queue employs two ida machines (`ida7` and `ida25`), each with 20 CPU cores and 128 GB RAM.
+As is typical for grid computing, all submitted jobs are sorted into specific [queues](https://docs.metacentrum.cz/computing/queues-in-meta/) (mainly based on the amount of requested resources). The combination of the required resources and the current infrastructure load determines the delay between the job submission and the start of the calculation. Very demanding jobs can wait in the queue for several days before all the required resources are free. We will use a special queue `MetaSeminar` reserved for this course to avoid this delay. This queue employs two ida machines (`ida7` and `ida25`), each with 20 CPU cores and 128 GB RAM.
> [!IMPORTANT]
-> Each job submitted during this course needs to target this dedicated queue. As you will see later, interactive jobs will include a parameter `-q MetaSeminar` and batch jobs will include a line `#PBS -q MetaSeminar`. In both cases, the job scheduler [PBSPro](https://docs.metacentrum.cz/basics/concepts/#pbs-servers) will send jobs to this specified queue.
+> Each job submitted during this course needs to target this dedicated queue. As you will see later, interactive jobs will include a parameter `-q MetaSeminar` and batch jobs will include a line `#PBS -q MetaSeminar`. In both cases, the job scheduler [PBSPro](https://docs.metacentrum.cz/computing/concepts/#pbs-servers) will send jobs to this specified queue.
## Data and tools
@@ -66,6 +66,7 @@ The following data and software tools will be used during the course:
- Illumina paired-end reads (NCBI SRA accession number: [SRX20115911](https://www.ncbi.nlm.nih.gov/sra/SRX20115911[accn])).
- Oxford Nanopore reads (NCBI SRA accession number: [SRX20115912](https://www.ncbi.nlm.nih.gov/sra/SRX20115912[accn])).
- [NCBI SRA Toolkit](https://github.com/ncbi/sra-tools) for downloading sequencing data.
+ - [FastQC](https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) for quality control of Illumina reads
-
## Useful links
@@ -83,10 +84,10 @@ The following data and software tools will be used during the course:
Like most computing/data centres, MetaCentrum nodes run exclusively on Linux (mainly [Debian](https://www.debian.org/)) and are controlled via the command line. Linux is preferred for its stability, security, speed, adaptability, and compatibility. Additionally, software tools for life sciences are primarily designed and optimised for Linux.
-We will use one of the login servers known as [frontend](https://docs.metacentrum.cz/basics/concepts/#frontends-storages-homes) for logging in. Frontend servers are accessible via SSH protocol and serve as a main gateway for the entire infrastructure.
+We will use one of the login servers known as [frontend](https://docs.metacentrum.cz/computing/frontends/) for logging in. Frontend servers are accessible via SSH protocol and serve as a main gateway for the entire infrastructure.
> [!WARNING]
-> Frontend servers are virtual machines with limited computational power and primarily serve for basic data inspection and manipulation, preparation of the shell scripts for batch jobs, short compilations, etc. Please do not use them for long and/or demanding calculations (rather, use an [interactive job](https://docs.metacentrum.cz/basics/jobs/#interactive-job)).
+> Frontend servers are virtual machines with limited computational power and primarily serve for basic data inspection and manipulation, preparation of the shell scripts for batch jobs, short compilations, etc. Please do not use them for long and/or demanding calculations (rather, use an [interactive job](https://docs.metacentrum.cz/computing/run-basic-job/#interactive-job)).
> [!NOTE]
> MetaCentrum can be accessed worldwide. We do not apply any geoblocking.
@@ -95,19 +96,19 @@ The following diagram shows the frontend servers' position (labelled as **Login
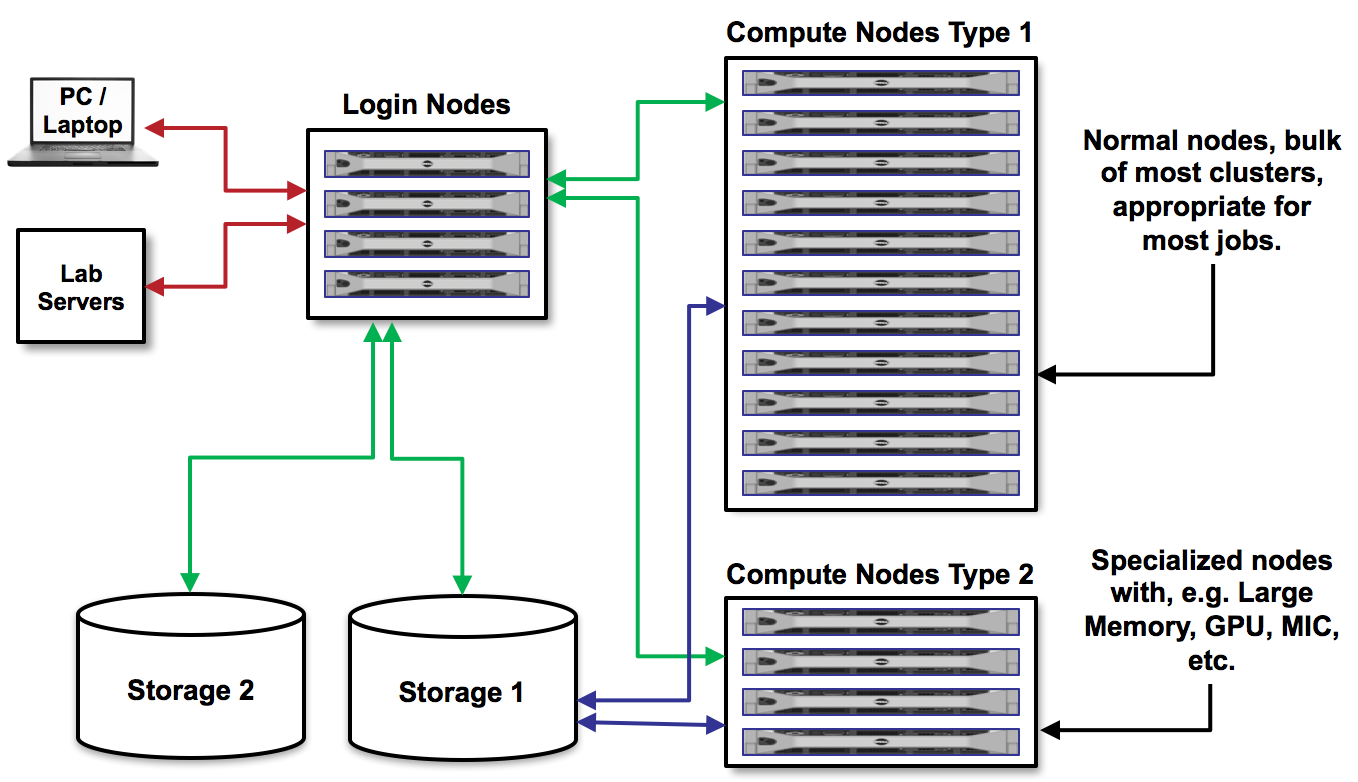
-In this tutorial, we will use frontend `skirit` with an address `skirit.metacentrum.cz` for logging in. Skirit frontend runs on Debian 11 and has a home directory mounted on the storage `brno2` (accessible as `/storage/brno2/home/$USER/`).
+In this tutorial, we will use frontend `nympha` with an address `nympha.metacentrum.cz` for logging in. Nympha frontend runs on Debian 11 and has a home directory mounted on the storage `plzen1` (accessible as `/storage/plzen1/home/$USER/`).
> [!IMPORTANT]
> MetaCentrum for log in does not fully support traditional authentication with SSH keys.
-> MetaCentum uses the [Kerberos](https://docs.metacentrum.cz/advanced/kerberos/) system for authentication, which requires a **username** and **password**.
+> MetaCentum uses the [Kerberos](https://docs.metacentrum.cz/access/kerberos/) system for authentication, which requires a **username** and **password**.
> [!TIP]
-> You can [install and configure Kerberos](https://docs.metacentrum.cz/advanced/kerberos/#install-kerberos) on your personal computer (available for all operating systems). It allows you to generate a local Kerberos ticket with a lifetime of up to 24 hours and log in to Metacentrum nodes without typing a password for this period.
+> You can [install and configure Kerberos](https://docs.metacentrum.cz/access/kerberos/#install-kerberos) on your personal computer (available for all operating systems). It allows you to generate a local Kerberos ticket with a lifetime of up to 24 hours and log in to Metacentrum nodes without typing a password for this period.
Windows users can use (for example) an SSH client PuTTY (as described [here](https://docs.metacentrum.cz/software/graphical-access/#connect-with-putty-in-windows)). CLI users can open their terminals and type the following command (**replace a string `user_name` with your actual MetaCentrum username**) and then your password.
```shell
-ssh user_name@skirit.metacentrum.cz
+ssh user_name@nympha.metacentrum.cz
```
> [!NOTE]
> No characters appear during the password typing in the terminal. This is a standard security behaviour.
@@ -189,9 +190,9 @@ You can execute the `ls` or `ls -lh` command again (depending on your preference
Expand this section and compare results with me.
```shell
-(BULLSEYE)vorel@skirit:~$ ls
+(BULLSEYE)vorel@nympha:~$ ls
test_directory test_file.txt
-(BULLSEYE)vorel@skirit:~$ ls -lh
+(BULLSEYE)vorel@nympha:~$ ls -lh
total 512
drwxr-xr-x 2 vorel meta 4.0K Nov 28 15:21 test_directory
-rw-r--r-- 1 vorel meta 0 Nov 28 15:25 test_file.txt
@@ -224,7 +225,7 @@ cat test_file.txt
Finally, we can try to rename the file `test_file.txt` to `test_file_renamed.txt` and move it into the directory `test_directory`.
> [!NOTE]
-> Remember that the `mv` command can be used for renaming and also for moving files and folders.
+> Remember, the `mv` command can rename and move files and folders.
```shell
mv test_file.txt test_file_renamed.txt
@@ -233,7 +234,7 @@ mv test_file.txt test_file_renamed.txt
mv test_file_renamed.txt test_directory
```
-And in the last step of this chapter, we can check whether the renamed file `test_file_renamed.txt` was moved to the correct position, and if yes, we can delete folder `test_directory` with its content because we will have no further use for it.
+In the last step of this chapter, we can check whether the renamed file `test_file_renamed.txt` was moved to the correct position, and if yes, we can delete folder `test_directory` with its content because we will have no further use for it.
```shell
ls test_directory
@@ -244,11 +245,87 @@ rm -r test_directory
# System of software modules
+Software tools available in MetaCentrum are accessible as [environment modules](https://modules.readthedocs.io/en/stable/index.html#). The system of modules is a concept simplifying the use of different software in a precise and controlled manner. Each program (in each version, eventually with additional modifications) is prepared as an individual module that must be activated (loaded) before use. The activation modifies the user's environment and sets everything necessary (especially variables $PATH and $LD_LIBRARY_PATH and loads dependencies) for the program's run.
-# Download raw data from NCBI SRA
+> [!TIP]
+> How to work with MetaCentrum modules is described in detail [here](https://docs.metacentrum.cz/software/modules/).
+> [!NOTE]
+> MetaCentrum provides a few thousand modules. With this number of modules, manual control is no longer possible, and sometimes, not everything works without problems. If you encounter any malfunction, please let us know at meta@cesnet.cz.
+We can execute a few commands and discuss how the module system works. Let's try to load a module for [BLAST+](https://blast.ncbi.nlm.nih.gov/doc/blast-help/downloadblastdata.html#downloadblastdata).
+```shell
+module ava blast
+```
+```shell
+module ava *blast*
+```
+```shell
+module ava blast-plus/
+```
+```shell
+module add blast-plus/2.12.0-gcc
+```
+```shell
+module list
+```
+```shell
+module rm blast-plus
+```
+```shell
+module list
+```
+> [!WARNING]
+> Remember that each activated module somehow modifies your environment. Loading many modules in one session can lead to conflicts and, as a result, to the non-functionality of some applications. The module that can be in conflict with others can be reliably limited only to defined parts of the job.
+
+```shell
+( module add module_name_1
+programme ...
+)
+# After the right-round bracket, the used module is inactive, and all environmental modifications are suppressed.
+```
+
+# Raw reads and quality control
+
+We will start this hands-on course by downloading the raw sequencing data from the [NCBI Sequence Read Archive](https://www.ncbi.nlm.nih.gov/sra), followed by a quality check.
+
+The methodology will include:
+- starting the interactive job and moving to the [scratch directory](https://docs.metacentrum.cz/computing/scratch-storages/).
+- downloading the raw reads.
+- quality control of Illumina and Oxford Nanopore raw reads.
+- a visual assessment of graphs in [OnDemand service](https://docs.metacentrum.cz/software/ondemand/).
+
+> [!IMPORTANT]
+> Scratch storage is a storage for temporary files for running jobs. This storage should be used only during computations and should be freed immediately after your job ends. The location of the scratch directory is defined by a system variable `SCRATCHDIR`.
+
+> [!TIP]
+> OnDemand is a service that enables users to access Metacentrum via a web browser. OnDemand allows access to files and directories using a graphical file manager, running graphical applications, or using a traditional terminal.
+
+First of all, we submit an interactive job. The meaning of individual parts of the command is explained below.
+
+```shell
+qsub -I -l select=1:ncpus=2:mem=10gb:scratch_local=20gb -l walltime=2:00:00 -q MetaSeminar
+```
+
+| Parameter | Action|
+| ------------- | -------------|
+| `qsub` | Command that submits jobs. |
+| `-I` | Declares that the job is to be run interactively. |
+| `select=1` | Reserves resources on one physical node. |
+| `ncpus=2` | Reserves two processors. |
+| `mem=10gb` | Reserves 10 GB of RAM. |
+| `scratch_local=20gb` | Reserves 20 GB of disk space on scratch. |
+| `walltime=2:00:00` | Reserves two hours for the job. |
+| `-q MetaSeminar` | Submits job into queue MetaSeminar. |
+
+After starting the job, go to the scratch directory, defined as the variable SCRATCHDIR.
+
+```shell
+cd $SCRATCHDIR
+```
+> [!IMPORTANT]
+> Variable SCRATCHDIR (`$SCRATCHDIR`) is automatically set for each job. Always use `$SCRATCHDIR` in each job. The real path to the scratch directory is unknown before the start of the job because contains the job number. For example `/scratch/user_name/job_123456789.meta-pbs.metacentrum.cz`.

