diff --git a/.gitignore b/.gitignore
index 0a764a4..a414392 100644
--- a/.gitignore
+++ b/.gitignore
@@ -1 +1,3 @@
+.DS_Store
+site/
env
diff --git a/docs/community/team.md b/docs/community/team.md
index cba59ae..2081507 100644
--- a/docs/community/team.md
+++ b/docs/community/team.md
@@ -1,15 +1,15 @@
## Team
-**Project Leader(s):**
+**Project Leader(s):**
[Stefano Moia](https://github.com/smoia) - Postdoc researcher at the École Polytechnique Fédérale de Lausanne
**Contributors:**
Thanks goes to these wonderful people for all their help (see [emoji key](https://allcontributors.org/docs/en/emoji-key))!
- [Phys2bids Contributors](https://github.com/physiopy/phys2bids#contributors-)
-- [Peakdet Contributors](https:https://hackmd.io/-M7a4dusTx6WBdN5WNaamg?both#//github.com/physiopy/peakdet#contributors-)
+- [Prep4Phys Contributors](https://github.com/physiopy/prep4phys#contributors-)
- [Phys2denoise Contributors](https://github.com/physiopy/phys2denoise#contributors-)
-This could be you! We welcome all skill levels with an interest in physiological data collection. Join a meeting and figure out how you can get involved, or check out the contributions guide.
+This could be you! We welcome all skill levels with an interest in physiological data collection. Join a meeting and figure out how you can get involved, or check out the contributions guide.
This project follows the [all-contributors](https://github.com/all-contributors/all-contributors) specification, meaning that contributions of any kind are recognized as such.
@@ -33,12 +33,12 @@ Thanks goes to these wonderful people ([emoji key](https://allcontributors.org/d

Vicente Ferrer
🐛 💻 📖 👀 ⚠️ |
- 
Sarah Goodale
📖🧑🏫📋 |
+ 
Sarah Goodale
📖🧑🏫📋 |

Yaroslav Halchenko
🚇 |

Soichi Hayashi
🐛 |

Vittorio Iacovella
💻 |
-
+

François Lespinasse
💻 ⚠️ |

Neville Magielse
📖 |

Ross Markello
🐛 💻 🖋 🤔 🚇 👀 ⚠️ 🧑🏫 |
diff --git a/docs/contributors-guide/getting-started.md b/docs/contributors-guide/getting-started.md
index 3fd0282..e8953bf 100644
--- a/docs/contributors-guide/getting-started.md
+++ b/docs/contributors-guide/getting-started.md
@@ -9,7 +9,7 @@ up some guidelines for contributions - to help you get involved ASAP! If
you lack knowledge in python development / github use / physiological
data handling, don't be scared! Try to jump in anyway. Most of the
original contributors learned these things exactly this way - jumping in
-and hoping to fall in the right way without breaking too many bones.
+and hoping to fall in the right way without breaking too many bones.
Contributions can be made in different ways, not only code! As we follow
the
@@ -34,7 +34,7 @@ Follow these three simple steps to get started:
Linux, Mac, and Windows developer installation
----------------------------------------------
-Note:
+Note:
The following instructions are provided assuming that python 3 is
**not** your default version of python. If it is, you might need to use
`pip` instead of `pip3`, although some OSs do adopt `pip3` anyway. If
diff --git a/docs/index.md b/docs/index.md
index 30e1d74..055dc8a 100644
--- a/docs/index.md
+++ b/docs/index.md
@@ -1,13 +1,13 @@
# Home
-Welcome to the main documentation site for the physiopy community and python packages! Please also check out our [Github Page](https://github.com/physiopy). For questions, you can always contact the project manager at [physiopy.community@gmail.com](mailto:physiopy.community@gmail.com).
+Welcome to the main documentation site for the physiopy community and python packages! Please also check out our [Github Page](https://github.com/physiopy). For questions, you can always contact the project manager at [physiopy.community@gmail.com](mailto:physiopy.community@gmail.com).
## Who are we?
-We are the physiopy development team consisting of an international group of volunteers. You can learn more about us and how to connect with us by visiting the [*Community*](/community) page. Contributions are also very much welcomed! Check out the [*Contributor Guide*](community/contributor-guide/) tab.
+We are the physiopy development team consisting of an international group of volunteers. You can learn more about us and how to connect with us by visiting the [*Community*](/community) page. Contributions are also very much welcomed! Check out the [*Contributor Guide*](/contributors-guide) tab.
## What is the importance of physiological data collection and its impact on MRI?
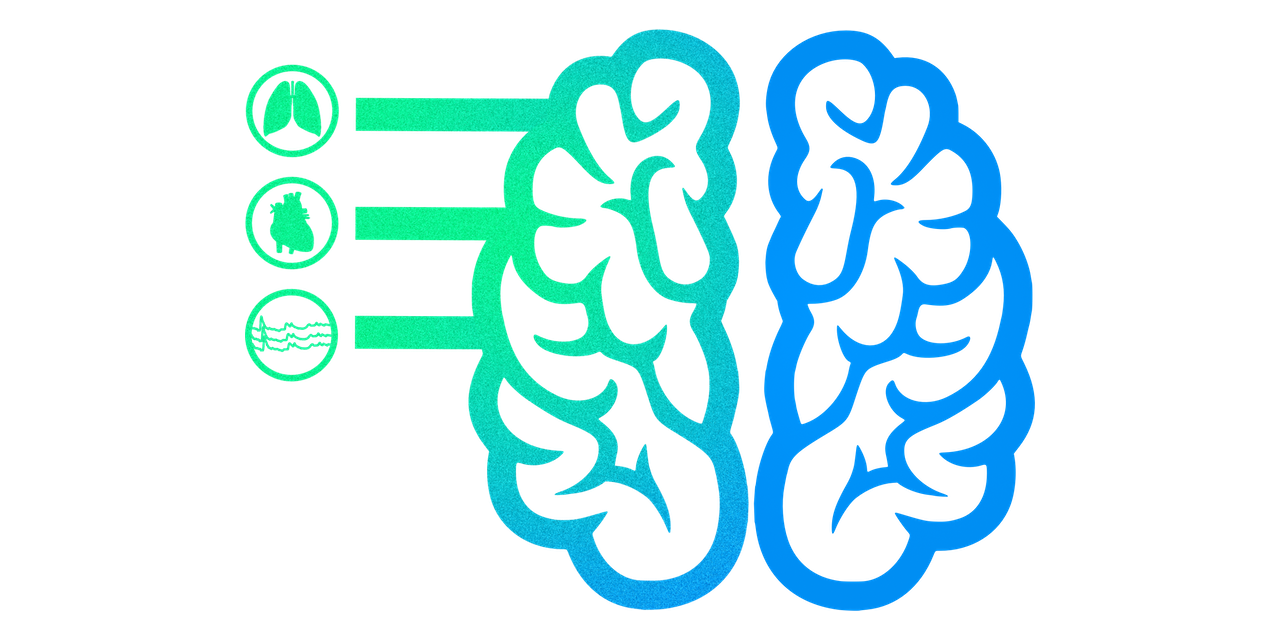
-Physiological data provides a rich representation of a research subject with respect to their bodily information (i.e., heart rate, respiratory rate and depth of breathing, skin conductance, etc.). Examining physiological activity helps us to understand the perception and embodied experience of cognition, emotion, motivation, and more. Additionaly, physiological data is a key component in understanding physiological sources of signal variance in fMRI data. Collecting these data helps to provide more accurate models of fMRI time-series. It also provides a real-time method to monitor subjects during scanning. See the [*Community Practices* documentation](https://physiopy-community-guidelines.readthedocs.io/en/latest/index.html) for recommendations from key personnel in the field on physiological data collection and analysis.
+Physiological data provides a rich representation of a research subject with respect to their bodily information (i.e., heart rate, respiratory rate and depth of breathing, skin conductance, etc.). Examining physiological activity helps us to understand the perception and embodied experience of cognition, emotion, motivation, and more. Additionaly, physiological data is a key component in understanding physiological sources of signal variance in fMRI data. Collecting these data helps to provide more accurate models of fMRI time-series. It also provides a real-time method to monitor subjects during scanning. See the [*Community Practices* documentation](https://physiopy-community-guidelines.readthedocs.io/en/latest/index.html) for recommendations from key personnel in the field on physiological data collection and analysis.
diff --git a/docs/libraries/peakdet.md b/docs/libraries/peakdet.md
deleted file mode 100644
index ca7c4f9..0000000
--- a/docs/libraries/peakdet.md
+++ /dev/null
@@ -1,17 +0,0 @@
-# Peakdet
-### A toolbox for physiological peak detection analyses
-
-This package `peakdet` is designed for use in the reproducible processing and analysis of physiological data, like those collected from respiratory belts, pulse photoplethysmography, or electrocardiogram (ECG/EKG) monitors. For github repository, follow this [link](https://github.com/physiopy/peakdet)
-
-### Overview
-Physiological data are messy and prone to artifact (e.g., movement in respiration and pulse, ectopic beats in ECG). Despite leaps and bounds in recent algorithms for processing these data there still exists a need for manual inspection to ensure such artifacts have been appropriately removed. Because of this manual intervention step, understanding exactly what happened to go from "raw" data to "analysis-ready" data can often be difficult or impossible.
-
-This toolbox, `peakdet`, aims to provide a set of tools that will work with a variety of input data to reproducibly generate manually-corrected, analysis- ready physiological data. If you'd like more information about the package, including how to install it and some example instructions on its use, check out our [documentation](https://peakdet.readthedocs.io/en/latest/)
-
-### Installation and User Guide
-Follow this for more information on [installation and setup](https://peakdet.readthedocs.io/en/latest/index.html) of `peakdet` as well as ways to get involved in the development and testing of this package.
-
-The [user guide](https://peakdet.readthedocs.io/en/latest/usage.html) also provided at the link above discusses the reproducibility of loading and saving data, as well as, the processing and editing of the physiological data.
-
-### License Information
-This codebase is licensed under the Apache License, Version 2.0. The full license can be found in the [LICENSE](https://github.com/physiopy/peakdet/blob/master/LICENSE) file in the peakdet distribution. You may also obtain a copy of the license [here](http://www.apache.org/licenses/LICENSE-2.0).
diff --git a/docs/libraries/prep4phys.md b/docs/libraries/prep4phys.md
new file mode 100644
index 0000000..0fba8f4
--- /dev/null
+++ b/docs/libraries/prep4phys.md
@@ -0,0 +1,23 @@
+# Prep4Phys
+
+**Previously named Peakdet**
+
+### A toolbox for physiological peak detection analyses
+
+This package `prep4phys` is designed for use in the reproducible processing and analysis of physiological data, like those collected from respiratory belts, pulse photoplethysmography, or electrocardiogram (ECG/EKG) monitors. For github repository, follow this [link](https://github.com/physiopy/prep4phys)
+
+### Overview
+
+Physiological data are messy and prone to artifact (e.g., movement in respiration and pulse, ectopic beats in ECG). Despite leaps and bounds in recent algorithms for processing these data there still exists a need for manual inspection to ensure such artifacts have been appropriately removed. Because of this manual intervention step, understanding exactly what happened to go from "raw" data to "analysis-ready" data can often be difficult or impossible.
+
+This toolbox, `prep4phys`, aims to provide a set of tools that will work with a variety of input data to reproducibly generate manually-corrected, analysis- ready physiological data. If you'd like more information about the package, including how to install it and some example instructions on its use, check out our [documentation](https://prep4phys.readthedocs.io/en/latest/)
+
+### Installation and User Guide
+
+Follow this for more information on [installation and setup](https://prep4phys.readthedocs.io/en/latest/index.html) of `prep4phys` as well as ways to get involved in the development and testing of this package.
+
+The [user guide](https://prep4phys.readthedocs.io/en/latest/usage.html) also provided at the link above discusses the reproducibility of loading and saving data, as well as, the processing and editing of the physiological data.
+
+### License Information
+
+This codebase is licensed under the Apache License, Version 2.0. The full license can be found in the [LICENSE](https://github.com/physiopy/prep4phys/blob/master/LICENSE) file in the prep4phys distribution. You may also obtain a copy of the license [here](http://www.apache.org/licenses/LICENSE-2.0).
diff --git a/mkdocs.yml b/mkdocs.yml
index b0ed01d..3451dbb 100644
--- a/mkdocs.yml
+++ b/mkdocs.yml
@@ -23,7 +23,7 @@ theme:
# Palette toggle for light mode
- scheme: default
toggle:
- icon: material/brightness-7
+ icon: material/brightness-7
name: Switch to dark mode
# Palette toggle for dark mode
@@ -38,7 +38,7 @@ nav:
- Libraries:
- phys2bids: libraries/phys2bids.md
- phys2denoise: libraries/phys2denoise.md
- - peakdet: libraries/peakdet.md
+ - prep4phys: libraries/prep4phys.md
- Contributor Guide:
- Welcome: contributors-guide/index.md
- Recognizing contributions: contributors-guide/recognition.md
@@ -58,7 +58,7 @@ nav:
- Welcome: contributors-guide/documentation/index.md
- Building documentation: contributors-guide/documentation/building.md
- Adding to Community Practices: contributors-guide/documentation/community-practices.md
- - Community Practices:
+ - Community Practices:
- Community Practices: https://physiopy-community-guidelines.readthedocs.io/en/latest/index.html
- Community:
- Welcome: community/index.md
@@ -69,6 +69,6 @@ nav:
- Tutorials:
- OHBM 2023 Tutorials: ohbm23_tutorials.md
- ReproNim 2024 Tutorials: repronim24_tutorials.md
-
+
