In life science, study of DNA is an important factor of understanding about organisms. DNA carries most of the genetic instructions of development, functioning, and reproduction in all organisms. Nowadays, with development of sequencing technologies, we can easily read a DNA sequence
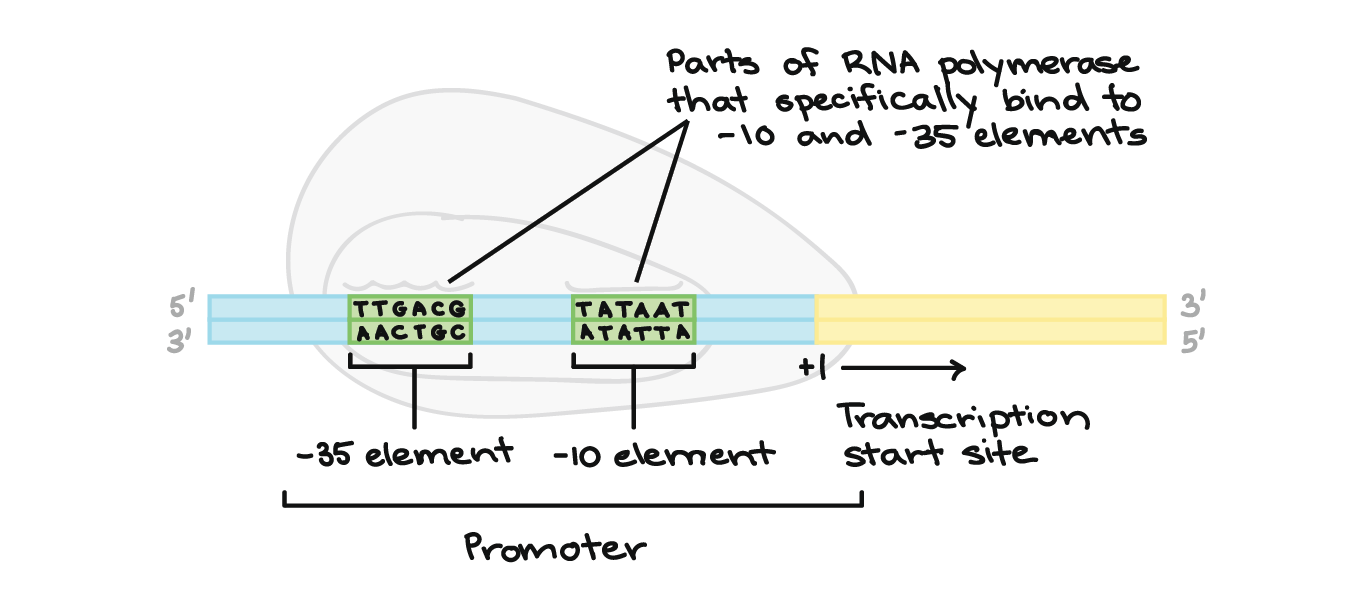
In this project , I created a model to classify DNA sequences of E.coli bacteria into two classes : Promoters/no promoters . A promoter is a region of DNA where transcription of a gene is initiated. Promoters are a vital component of expression vectors because they control the binding of RNA polymerase to DNA. RNA polymerase transcribes DNA to mRNA which is ultimately translated into a functional protein. Thus the promoter region controls when and where in the organism your gene of interest is expressed.
After the process of visualising and pre-processing the dataset provided from the Department of Molecular Biology and Biochemistry, results -referring to the F1-Score metric- showed that models tended to perform better with the linear SVM algorithm .
About the DATASET:
1.Title of Database: E. coli promoter gene sequences (DNA) with associated imperfect domain theory
2.Number of Instances: 106
-
Number of Attributes: 59 -- class (positive or negative) -- instance name -- 57 sequential nucleotide ("base-pair") positions
-
Attribute information:
Description:
1 One of {+/-}, indicating the class ("+" = promoter). 2 The instance name (non-promoters named by position in the 1500-long nucleotide sequence provided by T. Record). 3-59 The remaining 57 fields are the sequence, starting at position -50 (p-50) and ending at position +7 (p7). Each of these fields is filled by one of {a, g, t, c}.
-- Statistics for numeric domains: No numeric features used.
-- Statistics for non-numeric domains:
Frequencies: Promoters Non-Promoters
A 27.7% 24.4%
G 20.0% 25.4%
T 30.2% 26.5%
C 22.1% 23.7%
-
Missing Attribute Values: none
-
Class Distribution: 50% (53 positive instances, 53 negative instances)