This repository has been archived by the owner on Jul 8, 2020. It is now read-only.
-
Notifications
You must be signed in to change notification settings - Fork 0
Galaxy toolshed installation of the tool
asmariyaz23 edited this page Jul 18, 2018
·
6 revisions
- Git clone Galaxy by following instructions here.
- cd </path/to/galaxy>
- Configure Galaxy (config/galaxy.yml) with settings below:
- admin_users:
- conda_prefix: </valid/path/>
- conda_auto_install: true
- conda_auto_init: true
- Run Galaxy:
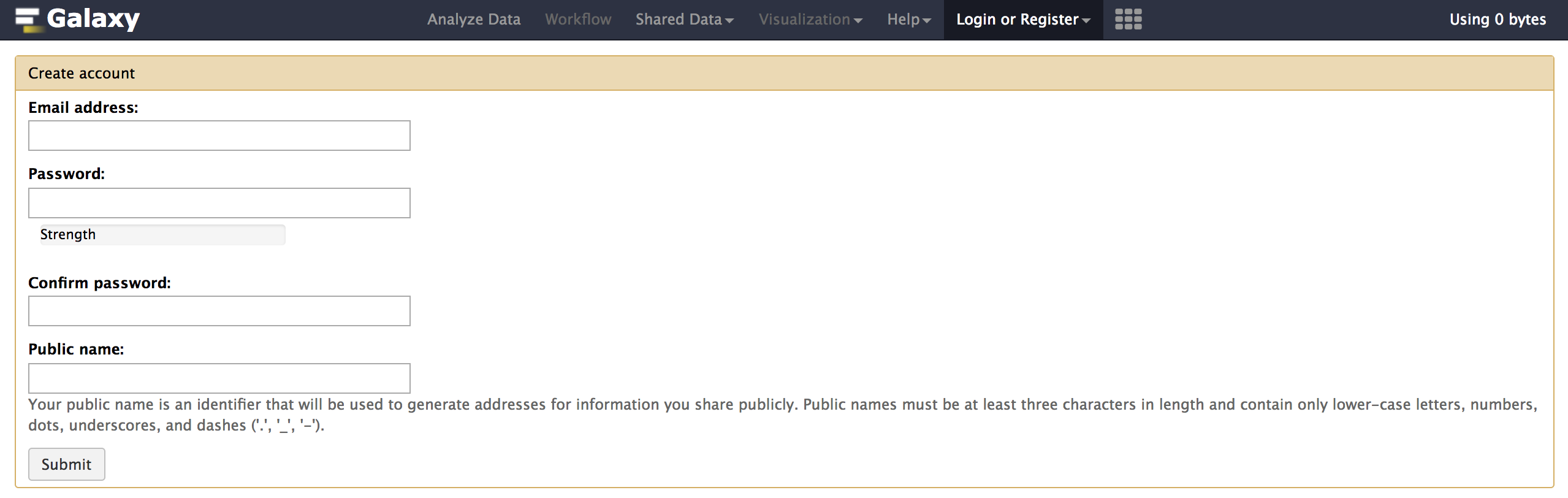
sh run.sh - Register and login with your admin email id used in config/galaxy.yml as below:
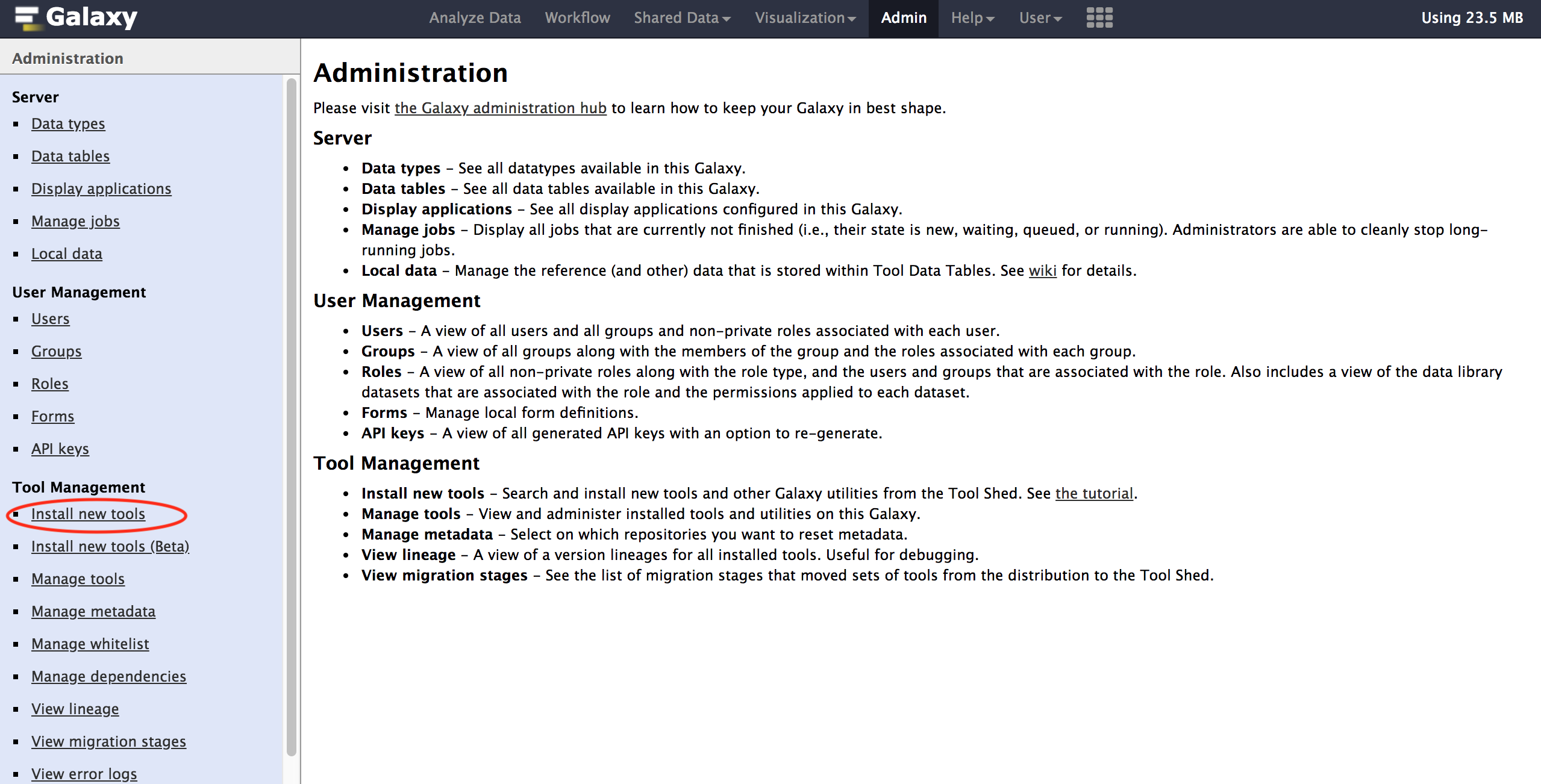
- Go to the Admin tab on the top panel
- Select Install new tools and select Galaxy Tool Shed
- Search for ctat_metagenomics and install it
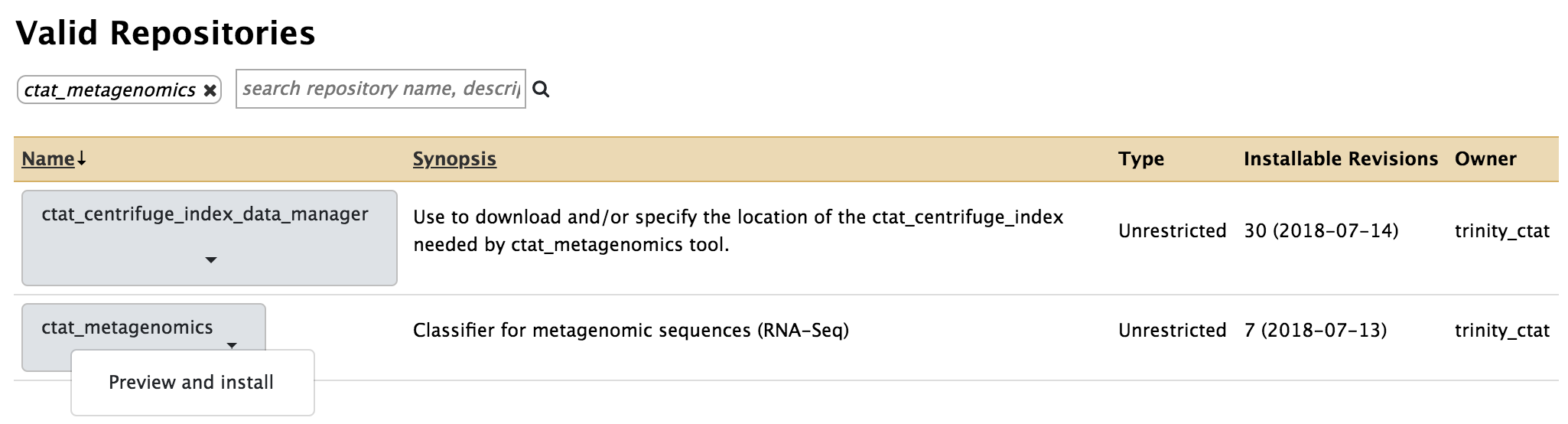
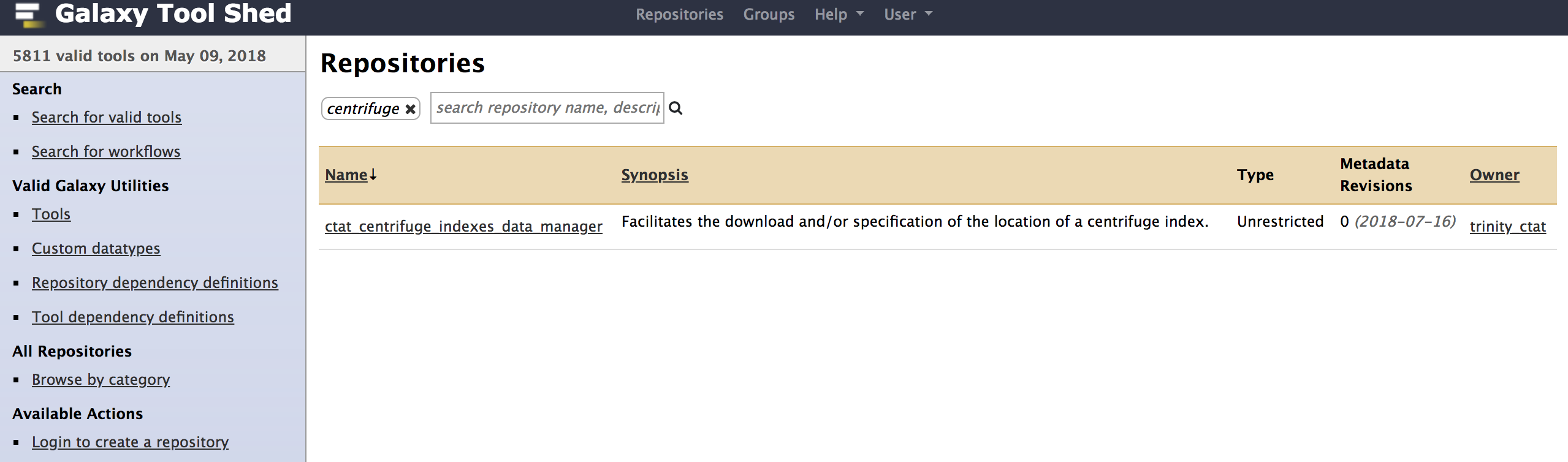
- Similarly install ctat_centrifuge_indexes_data_manager
- Next download to ctat_metagenomics data resource on your server/machine, click on Local Data (under Admin Tab). Under Data Manager Tools select CTAT metagenomics annotations Data Manager and fill the form as below (this may take a while):
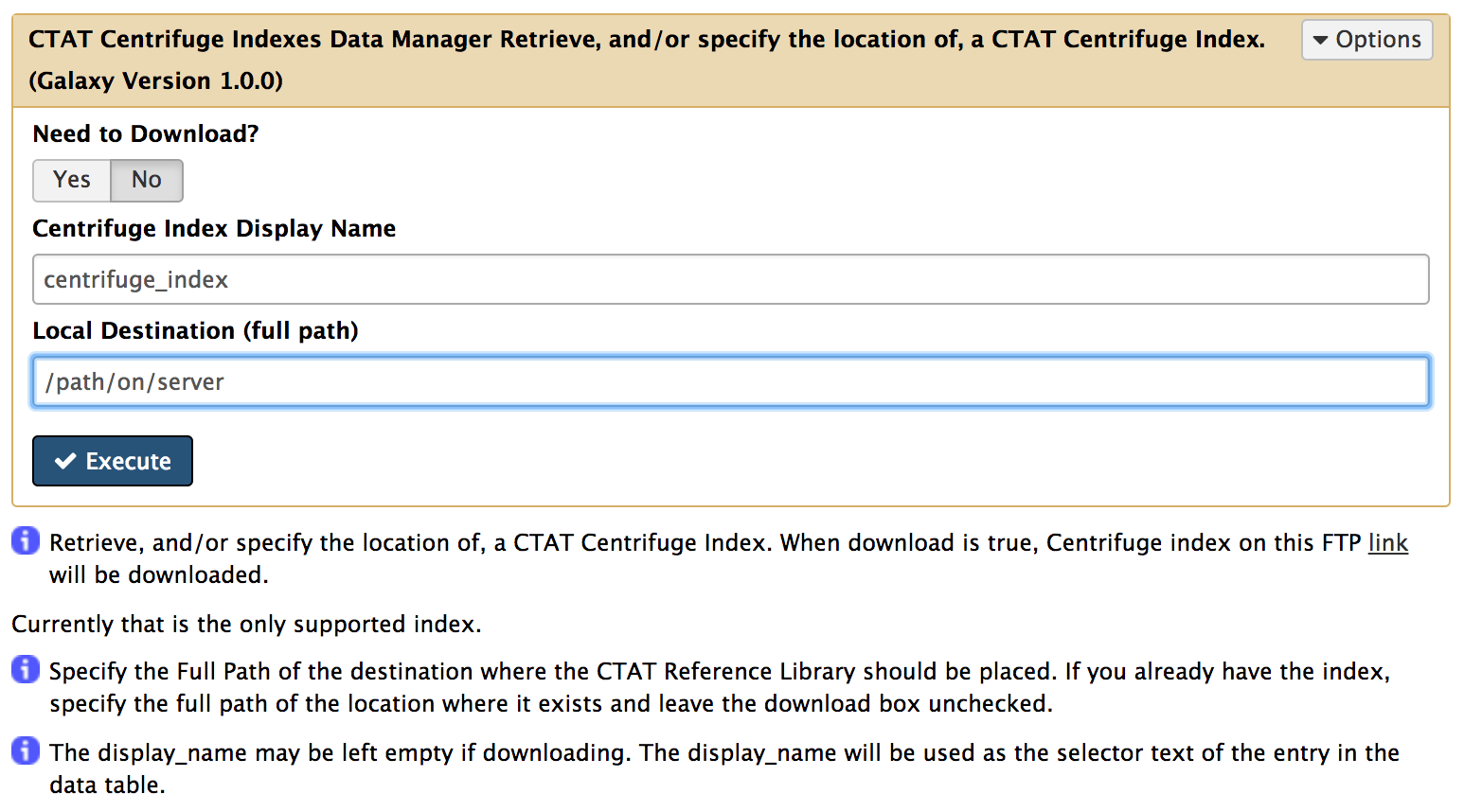
- If install and download was a success then upload fasta/fastq files using the Get Data tool. Some test data is available here.
- Run the ctat-metagenomics tool, leave all settings to default and select required genome.
Kim, D., Song, L., Breitwieser, F. P., & Salzberg, S. L. (2016). Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Research, 26(12), 1721–1729. https://doi.org/10.1101/gr.210641.116