-
Notifications
You must be signed in to change notification settings - Fork 4
masker example
The files used to perform this example can be found in bin/examples/
All example commands assume the subject and query files are in the same directory.
- Our genome file will be
LpKY97.fasta- This is a fungal genome which has been resolved to the chromosome level. It can also be downloaded here.
Our goal in this example is to mask all repetitive regions in the LpKY97 genome using FACET's masker module
To mask all repetitive regions in the LpKY97 genome, the following FACET command can be used:
FACET masker LpKY97.fasta -v
FACET will then run a self-BLAST and use the alignment information to mask repetitive regions in the genome. If you want more information on what constitutes a 'repetitive region', check this out.
Because we specified the verbose flag (-v), information about what FACET is doing will be printed to the stdout.
The end of that should look something like this:

Once FACET has finished (this may take a couple minutes depending on how large the genome is), a masked genome file will be output in the directory the program was called from. Lets take a peek at the head of the masked file:
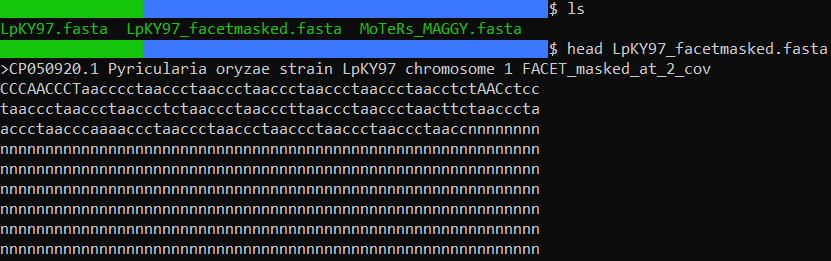
We can easily see that some bases at the very beginning of the genome have been masked with lowercase n's. Mission accomplished!