-
Notifications
You must be signed in to change notification settings - Fork 7
Gap filling feature
Lauren Coombe edited this page Apr 29, 2022
·
1 revision
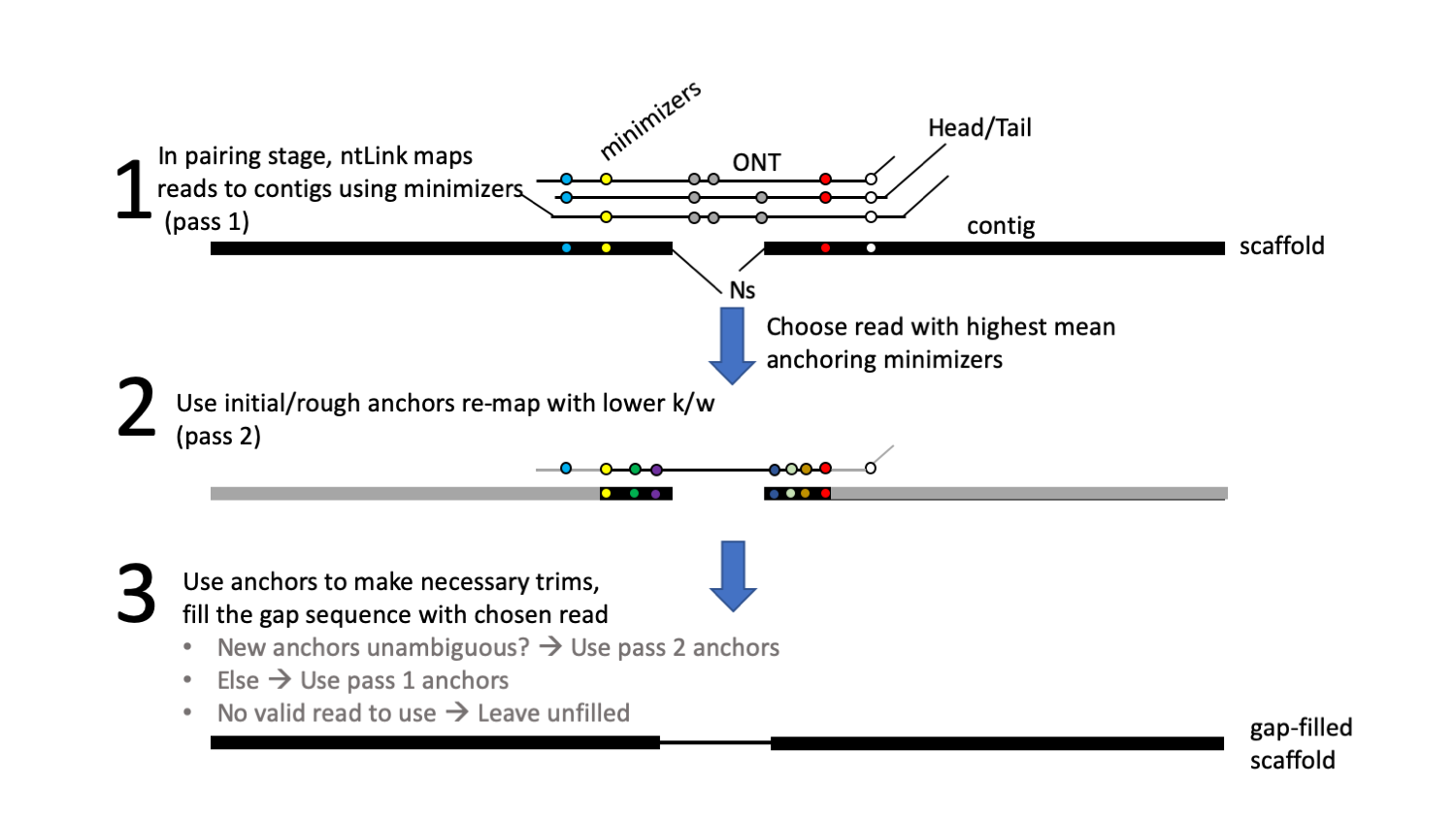
The ntLink gap_fill feature was introduced in ntLink v1.2.0, and provides functionality for filling ntLink-induced scaffold gaps with read sequence.
Most scaffolders will introduce ambiguous sequence (Ns) between joined sequences. If left unfilled, these regions of the assembled genome remain unknown and can interfere with various downstream analyses such as gene annotation.
This mode is enabled by adding the gap_fill target to the ntLink command. overlap=True is required when using the gap-filling feature.
Note that the gaps will be filled with raw read sequence, so subsequent polishing is a good idea.