-
Notifications
You must be signed in to change notification settings - Fork 1
MultipleReplicationExperiment
Running an experiment of the MultipleReplicationExperiment class.
The MultipleReplicationExperiment class can be used to run the same simulation experiment a number of times, using different streams of (pseudo)random numbers. This is commonly done in simulation studies to isolate the stochastic influence of random variables, which can have a large impact on the results of an experiment. The instructions in this section build on those on how to [DynamicShopExperiment run a job shop/flow shop experiment].
Just like the DynamicShopExperiment class, the MultipleReplicationExperiment class contains a number of parameters. However, we are only interested in setting the number of replications of the experiment (maxReplications) to 30 in this case. The only other parameter that we are required to set is baseExperiment, which refers to the underlying experiment of which we want to run several replications. Click on the “New” button, select DynamicShopExperiment and click on “OK”.
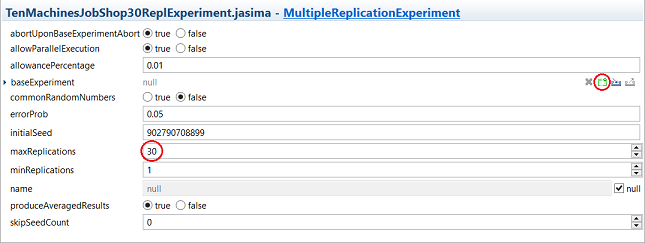
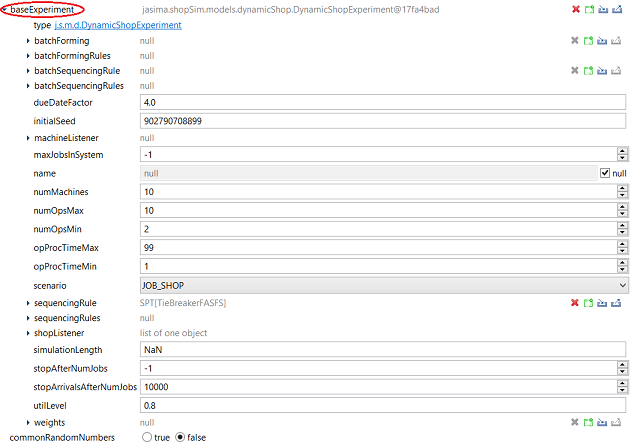
Next, click on baseExperiment to open the submenu of this parameter, which is identical to the main menu of the DynamicShopExperiment. We can set up this base experiment as described in the section on how to [DynamicShopExperiment run a job shop/flow shop experiment].
The experiment can be configured and run in the same way as an experiment of the DynamicShopExperiment class. The following section illustrates the basic structure of the resulting Excel output files. Since this is independent of the activated shop and machine listeners we only discuss one Excel file (generated with the default shop listener).
Once jasima® has completed all 30 replications, open the Excel file. You will notice that the spreadsheet consists of several worksheets.
 (screenshot showing the main experiment sheet, red circle to emphasise other sheets)
(screenshot showing the main experiment sheet, red circle to emphasise other sheets)
As in the case of the DynamicShopExperiment class, the “main experiment” worksheet consists of an upper section describing the parameter setting of the simulation experiment and a lower section showing the experimental results. In addition to the parameter values that are directly related to the replication experiment, the upper section also lists all parameter values of the base experiment, which are indicated by the prefix “baseExperiment”. Note that the lower section shows the results of the replication experiment (not the base experiment), i.e. single value measures such as the number of started jobs (“numJobsStarted”) are now summarised by a statistic, where the value in the “value/mean” column is the number of started jobs averaged over the 30 replications. For statistics such as flowtime, only the mean is shown (as indicated by the suffix “mean”), e.g. the value in the “min” column is the minimum mean flow time of jobs obtained in any of the 30 replications. Note also that the number of counted values (or means) for these measures is 30, i.e. equal to the number of replications.
(NOT SURE WHETHER THIS IS STILL IMPORTANT: It is also possible to assign an existing experiment to the baseExperiment property. To do so, click on the Load icon and change the file filter to 'Jasima experiments'. A copy of the experiment will be made, so changes in the original file are not visible in the outer experiment's file.)
Next, open the “sub-exp. overview” worksheet.
 (screenshot showing the sub-exp. overview sheet)
(screenshot showing the sub-exp. overview sheet)
This spreadsheet lists the parameter setting of each individual base experiment. Since experiments of the MultipleReplicationExperiment class simply replicate the base experiment a number of times, most parameter values are exactly the same for the individual base experiments. In fact, the only two parameters that take on a different value in every single base experiment (as indicated by the 30 in the “distinct values” column) are the initialSeed and name. The seed is a number that initialises the generator of the (pseudo)random number stream. The same seed will always yield the same random numbers (which is essential for the reproducibility of results), whereas a different seed generally corresponds to a different stream. Thus, the 30 distinct seed values indicate that each of the 30 replications is a different instance of the base experiment. Next, open the “sub-exp. value|mean” worksheet.
 (screenshot showing the sub-exp. overview sheet)
(screenshot showing the sub-exp. overview sheet)
This spreadsheet provides detailed information on the results of every replication of the base experiment, where each row relates to one replication. The first two columns identify an experiment by its seed and name, followed by a number of columns showing the results of the various measures (which depend on the activated listeners). For statistics such as flowtime, only the mean is shown in this worksheet. The other recorded statistical values, i.e. the minimum value, the maximum value, the standard deviation of values, the number of counted values, and the sum of all values, can be found in the subsequent worksheets that have exactly the same structure.