-
Notifications
You must be signed in to change notification settings - Fork 2
The mass spectrometry metadata editor
In order to edit or create a new template with Mass Spectrometry metadata that can be attach to an input file and an associated MS file (mgf or mzML), you have to use the MS metadata editor. Using it, some Mass Spectrometry metadata can be introduced in a set of forms and can be saved as a template for its use with further datasets.
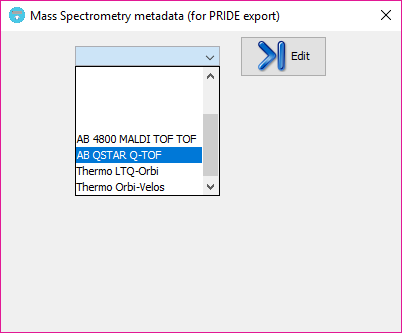
The metadata templates combo box will show any MS metadata information that the user has previously saved (or some example ones that comes by default with PACOM).
In order to edit or create a new template, just click on the 'Edit' button and wait a few seconds to the MS metadata editor to appear:
Once the MS metadata editor is open you will see the different sections of the metadata, based on the MIAPE Mass Spectrometry 1 specifications:
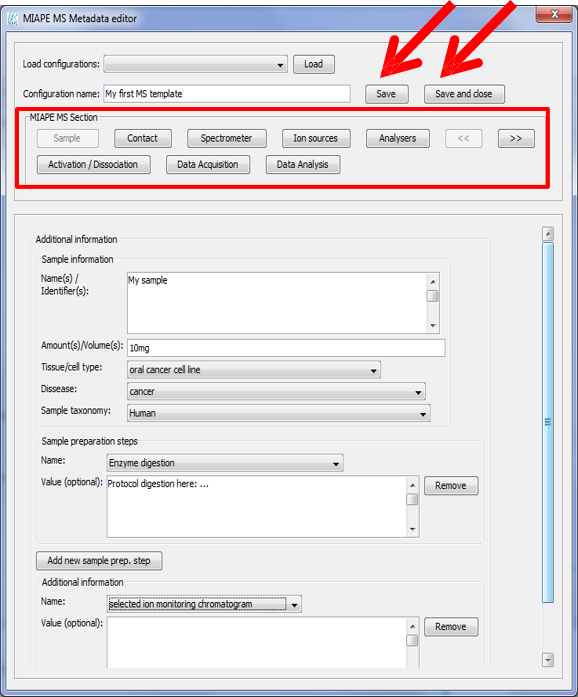
You will be able to jump to any of the metadata sections by clicking on the buttons in 'MIAPE MS Section'.
After filling the information that you want to include, click on 'Save' after assigning a name to the template. Click on 'Save and close' to save the template and close the MS metadata editor.
After closing the MS metadata editor, a new option will be available and already selected on the MS metadata combo box, and a summary of the metadata template will be shown just below:
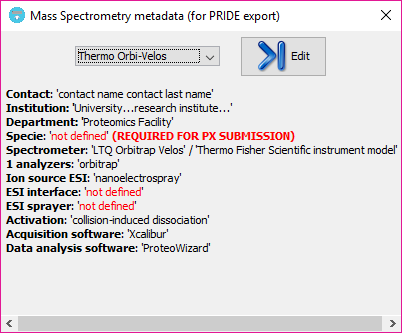
All MS metadata templates are stored in the system under the 'user_data\ms_metadata_templates' folder. If some information is mandatory for a ProteomeXchange submission is missing, the section will be highlighted with red color as:
Note: That information will be missed in the PRIDE XML that you can generate at the end of the workflow. However, you should take into account that a ProteomeXchange submission using PRIDE XML files into the PRIDE EBI repository is not recommended and you rather be use the standard mzIdentML for that. So, in summary, the tool offers a way to create PRIDE XML files that are MIAPE-compliant, but its use for a ProteomeXchange submission is now deprecated.
Contact person for any suggestion or error report:
Salvador Martínez-Bartolomé (salvador at scripps.edu)
Senior Research Associate
The Scripps Research Institute
10550 North Torrey Pines Road
La Jolla, CA 92037
Git-Hub profile